Abstract
Paris liiana sp. nov is a species of flowering herb of the genus Paris and widely distributed in the southwest of China. In this study, we sequenced the complete chloroplast (cp) genome of P. liiana sp. nov to investigate its phylogenetic relationship in genus Paris. The cp genome of P. liiana sp. nov was 163,860 bp in length, containing a large single-copy (LSC) region of 84,415 bp, a small single-copy (SSC) region of 12,947 bp, and a pair of inverted repeats (IRs) region of 33,249 bp. The overall GC content was 37.0%. The genome comprises of 135 genes, including 91 protein-coding genes, 37 tRNA genes, and 4 rRNA genes. Phylogenetic relationship analysis based on complete cp genome sequences exhibited that P. liiana sp. nov was most related to P. polyphylla var. yunnanensis.
Paris polyphylla var. yunnanensis is a perennial rhizomatous herb distributed in southwestern China and northern Myanmar (Li Citation1998). Studies have shown that Paris plant mainly contains steroidal saponins, triterpenoids, flavonoids and other chemical components, Steroidal saponins are the major active chemical constituents of Paris plants (Wang et al. Citation2015), including antitumor activity (Yan et al. Citation2009; Man et al. Citation2011), immuno-stimulating properties (Zhang et al. Citation2007), anthelmintic activity (Wang et al. Citation2010), antimicrobial activity (Qin et al. Citation2012), and protective effects on indomethacin-induced gastric mucosal lesions. It is also the material basis of some Chinese patent anticancer medicines, such as Gan-Fu-Le capsules, Bo-Er-Ning capsules, Lou-Lian capsules and so on (Wang et al. Citation2018). In the early stage of this study, we found some morphological differences among different samples of P. polyphylla var. yunnanensis, two phenotypes (“typical” and “high stem”). Paris polyphylla var. yunnanensis taxonomic rank has been in dispute. This study is about P. polyphylla var. yunnanensis (‘high stem’), which is described as P. liiana sp. nov. It shows significant morphological differences from ‘typical’ P. polyphylla var. yunnanensis, which include plant height, leaf-blade shape, length, and width, sepal shape, petal color and width, and color of fruit at maturity (Ji et al. Citation2020). But. they are the same species based on traditional classification. Ji et al. (Citation2020) proposed that P. liiana sp. nov might be a new species. Therefore, it is needed to accurately identify this species used the complete chloroplast genome of P. liiana sp. nov.
Fresh, healthy, and clean leaf materials of P. liiana sp. nov were collected from Gucheng District, Dali, Yunnan, China (25°69′14″'N, 100°16′25″E), the voucher specimen was collected and deposited at the Herbarium of Dali University (20191002B4). The total DNA was extracted using the DNeasy plant mini kit (QIAGEN), and next-generation sequencing was carried out by an Illumina NovaSeq system (Illumina, San Diego, CA, USA). About 7.9 Gb of raw data (52,505,044 reads) were assembled by NOVOPlasty (Park et al. Citation2019, Fan Citation2020), and the assembled cp genome was annotated by GeSeq with default sets (Tillich et al. Citation2017,Yang et al. Citation2020). The annotated cp genome was submitted to the GenBank with the accession number of MT857225.
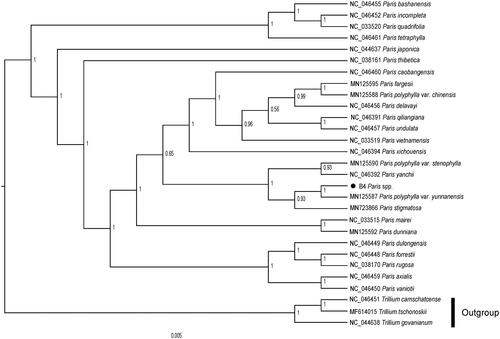
The cp genome of P. liiana sp. nov was 163,860 bp in length, containing a large single-copy (LSC) region of 84,415 bp, a small single-copy (SSC) region of 12,947 bp, and a pair of inverted repeats (IRs) region of 33,249 bp. The overall GC content was 37.0%. The genome comprises of 135 genes, including 91 protein-coding genes, 37 tRNA genes, and 4 rRNA genes. To confirm the phylogenetic position of P. liiana sp. nov, a total of 28 cp genome sequences were downloaded from the NCBI database. After using MAFFT V.7.149 for aligning (Kazutaka and Standley Citation2013), jModelTest v.2.1.7 (Darriba et al. Citation2012) was used to determine GTR + I + G as the best-fitting model for the chloroplast genomes. Then, Bayesian inference (BI) tree was performed by MrBayes v.3.2.6 (Fredrik et al. Citation2012), with Trillium tschonoskii (MF614015), Trillium camschatcense (NC_046451) and Trillium govanianum (NC_044638) as outgroups. The results of tree-building showed that P. liiana sp. nov was sister to P. polyphylla var. yunnanensis. It indicated that they possessed close phylogenetic relationships with each other (). However, Ji et al. (Citation2020) proposed that P. liiana sp. nov may be a new species (Ji et al. Citation2020). An in-depth study were needed to accurately verify this species with increased sampling of P. liiana sp. nov and P. polyphylla var. yunnanensis. The cp genome sequence of P. liiana sp. nov reported in this study may provide useful resources for the taxonomy and phylogeny of Paris genus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI GenBank database at (https://www.ncbi.nlm.nih.gov/nuccore/MT857225) with the accession number is MT857225, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Additional information
Funding
References
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772.
- Fan MLJWJ. 2020. The first complete chloroplast genome sequence of Paris polyphylla var. emeiensis, a rare and endangered species. Mitochondrial DNA B. 5(3):2172–2173.
- Fredrik R, Maxim T, Paul VDM, Ayres DL, Aaron D, Sebastian H, Bret L, Liang L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Ji Y, Liu C, Yang J, Jin L, Yang Z, Yang J-B. 2020. Ultra-barcoding discovers a cryptic species in Paris yunnanensis (Melanthiaceae), a medicinally important plant. Front Plant Sci. 11:411.
- Kazutaka K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li H. 1998. The genus Paris (Trilliaceae). Beijing: Science Press.
- Man S, Gao W, Zhang Y, Ma C, Yang L, Li Y. 2011. Paridis saponins inhibiting carcinoma growth and metastasis in vitro and in vivo. Arch Pharm Res. 1(34):43–50.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Park J, Choi YG, Yun N, Xi H, Min J, Kim Y, Oh S-H. 2019. The second complete chloroplast genome sequence of the Viburnum erosum (Adoxaceae) showed a low level of intra-species variations. Mitochondrial DNA A. 4(2):3278–3279.
- Qin XJ, Sun DJ, Ni W, Chen CX, Hua Y, He L, Liu HY. 2012. Steroidal saponins with antimicrobial activity from stems and leaves of Paris polyphylla var. yunnanensis. Steroids. 77(12):1242–1248.
- Wang GX, Han J, Zhao LW, Jiang DX, Liu YT, Liu XL. 2010. Anthelmintic activity of steroidal saponins from Paris polyphylla. Phytomedicine. 17(14):1102–1105.
- Wang YH, Niu HM, Zhang ZY, Xiang-Yang HU, Heng LI. 2015. Medicinal values and their chemical bases of Paris. China J Chin Mater Med. 5(40):833–839.
- Wang YH, Shi M, Niu HM, Yang J, Xia MY, Luo JF, Chen YJ, Zhou YP, Li H. 2018. Substituting one Paris for another? In vitro cytotoxic and in vivo antitumor activities of Paris forrestii, a substitute of Paris polyphylla var. yunnanensis. J Ethnopharmacol. 218:45–50.
- Yan LL, Zhang YJ, Gao WY, Man SL, Wang Y. 2009. In vitro and in vivo anticancer activity of steroid saponins of Paris polyphylla var. yunnanensis. Exp Hematol Oncol. 1(31):27.
- Yang K, Shang M, Jiang Y, Qian J, Duan B, Yang Y. 2020. The complete chloroplast genome of Rumex hastatus D. Don and its phylogenetic analysis. Mitochondrial DNA B. 5(2):1681–1682.
- Zhang XF, Cui Y, Huang JJ, Zhang YZ, Nie Z, Wang LF, Yan BZ, Tang YL, Liu Y. 2007. Immuno-stimulating properties of diosgenyl saponins isolated from Paris polyphylla. Bioorg Med Chem Lett. 17(9):2408–2413.