Abstract
Thermopsis turkestanica Gand. is a perennial herbaceous plant belonging to the genus Thermopsis, Leguminosae, and is mainly distributed in dry areas. Most of the species in this genus have high medicinal value. The complete chloroplast genome was reported in this study. The chloroplast genome with a total size of 149,551 bp consists of two inverted repeats (IRs, 24,159 bp) separated by a large single-copy region (LSC, 83,692 bp) and a small single-copy region (SSC, 17,541 bp). Further annotation revealed the chloroplast genome contains 110 genes, including 77 protein coding genes, 29 tRNA genes, and four rRNA genes. This information will be useful for study on the evolution and genetic diversity of Thermopsis turkestanica in the future.
Thermopsis turkestanica Gand. is a perennial herbaceous plant belonging to the genus Thermopsis, Leguminosae, and mainly distributed in the river valley and hillside of Xinjiang Tianshan Belt of China, it is also distributed in Mongolia, Altai, Kazakhstan, Uzbekistan, Turkmenistan, Kyrgyzstan, Tajikistan, and western Siberia of Russia. Most plants in this genus are frequently used as folk medicine in Northwest and Northeast China, such as the anti-cancer effect (Gao and Zhu Citation1999; Yildiz et al. Citation2020). In this study, to obtain the new insight into the phylogeny of T. turkestanica, we assembled and annotated the accurate chloroplast genome from sequenced data using Illumina HiSeq platform.
The materials of T. turkestanica in this study were collected from Aheqi County, Kizilsu Kirghiz Autonomous Prefecture, Xinjiang province of China (77°39.017′E, 40°49.949′N, 2464 m above sea level). The voucher specimen (TD-01531, Thermopsis turkestanica Gand.) was stored in the herbarium of Tarim University. The complete genomic DNA was extracted using CTAB method and sequenced using the Illumine HiSeq platform. First, we remove low quality sequences and joints from the raw data (SRR12880891), then quality control. The whole chloroplast genome was assembled using GetOrganelle (Jin et al. Citation2020). Then, the chloroplast gene structures were annotated using CPGAVAS2 (Shi et al. Citation2019) and PGA (Qu et al. Citation2019). The complete chloroplast genome was 149,551 bp (MW122835) and composed of two inverted repeats (IRs) of 24,159 bp each, which divide a large single-copy (LSC) region of 83,692 bp and a small single-copy (SSC) region of 17,541 bp, the average GC content was 36.41%. The chloroplast genomes encoded 110 functional genes, including 77 protein-coding genes, 29 tRNA genes, and four rRNA genes.
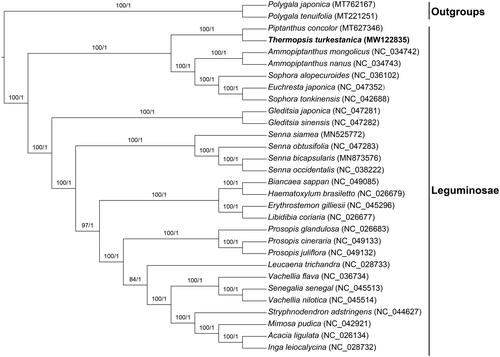
In our study, to explore the phylogenetic relationship of T. turkestanica within Leguminosae, additional 27 species from Leguminosae were studied. With the Polygala japonica and Polygala tenuifolia as the outgroups, the phylogenetic trees were built from the whole protein-coding gene matrix by maximum-likelihood (ML) and Bayesian inference (BI) (). The ML tree was generated using IQ-TREE (Nguyen et al. Citation2015) based on the best model of TVM + F+R4 and 1000 bootstrap replicates, and BI analysis was performed in MrBayes-3.2.7. This result showed that the analyzed T. turkestanica was closer to the species of Piptanthus concolor. This information will provide the basis for the study of Thermopsis turkestanica in the future.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW122835. The associated ‘BioProject’, ‘SRA’, and ‘Bio-Sample’ numbers are PRJNA670258, SRR12880891, and SAMN16491007, respectively.
Additional information
Funding
References
- Gao WY, Zhu DY. 1999. Advance in chemical studies of genus Thermopsis. Nat Prod Res Dev. 11(6):79–84.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Qu X-J, Moore MJ, Li D-Z, Yi T-S. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Yildiz M, Terzi H, Yildiz SH, Varol N, Erdoğan MÖ, Kasap M, Akçali N, Solak M. 2020. Proteomic analysis of the anti-cancer effect of various extracts of endemic Thermopsis turcica in human cervical cancer cells. Turk J Med Sci. 50.