ABTRACT
The common strain black carp (Cyprinus carpio var. baisenensis), known for its black skin is cultured in the integrated rice-agriculture system and non-escape property under torrential floods. The total mitogenome length of Cyprinus carpio var. baisenensis obtained in this study was 16,478bp, consisting of 13 protein-coding genes, 22 tRNA genes, 2rRNA genes (large and small), a light strand origin of replication, and one major non-coding region. By providing the complete mitochondrial genomes of Cyprinus carpio var. baisenensis, we will further understand the phylogenetic relationships within genus.
Introduction
The common carp (Cyprinus carpio) is a widespread freshwater fish originated in Eurasia. Over the years, the common carp has become one of the most important edible and popular aquarium fish, comprising a large number of strains worldwide because of its cultural history and domestication (Zhou et al. Citation2003). Common carp populations have the highest numbers of domesticated strains in china compared to other countries worldwide. These renowned strains include Huanghe carp (HH) Genome Wide Identified Main QTL Related to Growth Performance of Huanghe Carp New Strain (Cyprinus carpio hacmalopterus), Oujiang color carp (OJ), Hebao red carp (HB), Songpu mirror carp (SP), Xingguo red carp (XC) and Genome Wide Identified Main QTL Related to Growth Performances of Huanghe Carp New Strain (Cyprinus carpio hacmalopterus Temminck et Schlegel) (Dong et al. Citation2015; Su et al. Citation2018).
The common strain black carp (Cyprinus carpio var. baisenensis) from Guangxi has a history with the local Bourau people who prefer the black color. The sub-species color reflects the interaction between Bourau and their surrounding environment. This species is selected, geographically isolated and breeded, so there is no germplasm pollution. The common strain black carp is cultured in an integrated rice culture. This culture system helps the individuals to persist when there are floods. Supplementary Figure S1 shows the environment of the black carp and the pictures of the black carp. Complete mitochondrial genome sequencing can provide useful information for species conservation and identification (Li et al. Citation2016). Complete sequencing of mitochondrial genome also plays an important role in the study of Cyprinus carpio var. baisenensis phylogeography and genetic diversity of the population (Guan et al. Citation2018). mtDNA sequences analysis is one of the most effective methods available for population studies because it is simple and allows analyzing a large numbers of samples (Hilbish Citation1996; Nishida Citation1998). The common strain black carp was obtained from Guangxi. In this study, the complete mitochondrial genome of Cyprinus carpio var. baisenensis was sequenced using the Next-generation sequencing technology on Illumina NovaSeq. The assemble software used is GetOrganelle v1.6.2e (Jin et al. Citation2020). This annotation method was conducted using MITOS Webserver (Bernt et al. Citation2013). Total genomic DNA was extracted from the muscle of Cyprinus carpio var.baisenensis using a DNA extraction kit (Simgen, China) following the manufacturer’s instructions. Twelve primer sets were used in the PCR amplification, and the amplification was carried out in a total volume of 25 ml including: 60 ng total genomic DNA, 1 U Ex-Taq DNA polymerase, 2.0 mM MgCl2, 2 mM dNTP, and 10pM of each primers. The total mitogenome length of the common strain black carp obtained in this study was 16,478bp in the, which was deposited in GenBank with the accession number MT780875, under the Biosample number of SAMN16679851, Bioproject ID of PRJNA674914, SRA of SRR12996635 and SRP 291369 in NCBL (https://www.ncbi.nlm.nih.gov/). The mitogenomes of Cyprinus carpio var. baisenensis consisted of 13 protein coding genes, 22 tRNA genes, 2rRNA genes (large and small), origin of light (OL) strand of replication and one major non-coding region (D loop) (). The Cyprinus carpio var. baisenensis sequence protein coding total length is 11,415bp and most of these proteins and tRNA genes are encoded on the D-strand (Direct strand).). The total length of all tRNA genes was 1,563bp and rRNA total length was 2,586bp. All the PCGs start with ATG codon expect for COX1 that starts with GTC (). Seven protein coding genes have TAA as stop codon (NAD1, COX1, ATP6, COX3, NAD4L, NAD5, NAD6) while NAD2, ATP8, NAD3 have TAG as stop codon. OL, COX2, NAD4 and COB have AAA, CCT, ATT and CTT, respectively, as their stop codon ().
Table 1. Mitochondrial genome characteristics of Cyprinus carpio var.baisenensis.
The length of 22 tRNA genes varies from 67 bp (tRNACys) to 76 bp (tRNALeu and tRNALys). The two rRNA genes have nucleotide size of 163 bp respectively and are located in a position between tRNAPhe and tRNALeu, and separated by tRNAVal. The L-strand origin of replication (OL), it is located identified between the tRNAAsn and tRNACys and has a size of 32 bp. The control region (D-loop) with the nucleotides size of 806 bp is located between tRNAPro and tRNAPhe ().
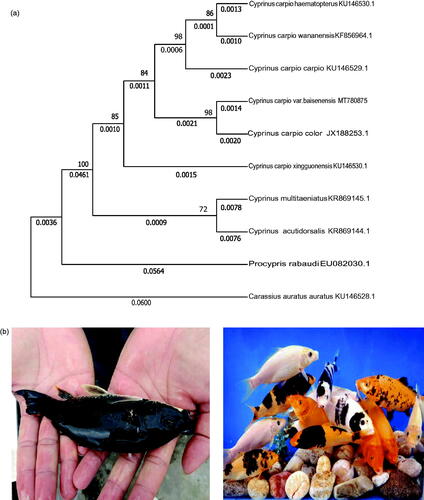
The phylogenetic position of Cyprinus carpio var. baisenensis was reconstructed based on the complete mitogenomes of11 species using maximum-likelihood (ML) methods and the MEGA X (Kumar et al. Citation2004). The phylogenetic tree of the common strain black carp shows that Cyprinus carpio var. baisenensis is the sister species of Cyprinus carpio color ().
Acknowledgments
The authors thank the students and staff of Aquatic Genetic Laboratory, FFRC for their kind assistance in the study. Thanks to Chenghui Wang from the Shanghai Ocean University for the Cyprinus carpio color picture.
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Availability of data and materials
The genome sequence data that support the findings of (Complete Mitochondrial Genomes (mtDNA) of Common Strain Black Carp (Cyprinus carpio var.baisenensis) are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT780875. The associated Bio-Project, SRR, SRP and Bio-Sample numbers are PRJNA674914, SRR12996635, SRP 291369, and SAMN16679851 respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Dong C, Xu J, Wang B, Feng J, Jeney Z, Sun X, Xu P. 2015. Phylogeny and evolution of multiple common carp (Cyprinus carpio L.) populations clarified by phylogenetic analysis based on complete mitochondrial genomes. Mar Biotechnol. 17(5):565–575.
- Guan M, Tan H, Fazhan H, Xie Z, Shi X, Zhang Y, Lin F, Ikhwanuddin M, Ma H. 2018. The whole mitochondrial genome and phylogenetic analysis of Lupocycloporus gracilimanus (Stimpson, 1858)(Decapoda, portunidae). Mitochondrial DNA Part B. 3(2):1244–1245.
- Hilbish TJ. 1996. Population genetics of marine species: the interaction of natural selection and historically differentiated populations. J Exp Mar Biol Ecol. 200(1–2):67–83.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31.
- Kumar S, Tamura K, Nei M. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5(2):150–163.
- Li G, Xu J, Wang H, Chen X, Xu Y, Zheng Y, Mao X, Yang S. 2016. Identification and analysis of the complete mitochondrial genome sequence of Eriocheir sinensis. Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):3923–3924.
- Nishida M. 1998. Methods of analysis of genetic population structure with mitochondrial DNA markers. Suisan Ikushu. 26:81–100.
- Su S, Li H, Du F, Zhang C, Li X, Jing X, Liu L, Li Z, Yang X, Xu P, et al. 2018. Combined QTL and genome scan analyses with the help of 2b-RAD identify growth-associated genetic markers in a new fast-growing carp strain. Front Genet. 9:592.
- Zhou JF, Wu QJ, Ye YZ, Tong JG. 2003. Genetic divergence between Cyprinus carpio carpio and Cyprinus carpio haematopterus as assessed by mitochondrial DNA analysis, with emphasis on origin of European domestic carp. Genetica. 119(1):93–97.