Abstract
Primula homogama F. H. Chen & C. M. Hu (Primulaceae) is endemic to the Emei Mountain of China. In this study, we characterized the complete chloroplast genome of P. homogama based on next-generation sequencing (NGS). The complete chloroplast genome of P. homogama was 154,677 bp in size with a typical quadripartite structure, containing a large single-copy (LSC) region of 85,299 bp and a small single-copy (SSC) region of 17,816 bp. These two regions were separated by a pair of inverted repeat regions (IRs), each of 25,781 bp. A total of 130 functional genes were encoded, consisted of 86 protein-coding genes (PCG), 36 tRNA genes, and eight ribosomal RNA (rRNA) genes.
Primula homogama F. H. Chen & C. M. Hu, belonging to sect. Souliei of Primulaceae, is a perennial herb species endemic to the Emei Mountain of China (Hu and Kelso Citation1996). Contrary to the distyly conditions for vast majority (92%) of Primula species (∼400–500), P. homogama was formerly recognized as monomorphic for stylar condition and homostyly with populations comprising a single floral form with anthers and stigmas close together within a single flower. These plants are generally self-compatible and predominantly selfing as a result of autonomous self-pollination (Ganders Citation1979). However, our preliminary observations of P. homogama has revealed the mixed populations containing both distylous and homostylous forms. Thus, the species represent a recent transition from distyly to homostyly (Zhong et al. Citation2019). The complete plastome could provide valuable genomic information for the phylogeny and population structure of this distylous-homostylous species.
Fresh leaves tissue of P. homogama were collected from a wild population (103°15′00″E, 29°30′00″N) in Hongya County, Meishan City, Sichuan Province. We isolated total genomic DNA following a modified CTAB protocol (Doyle Citation1991). The purified genomic DNA was sheared into c. 500 bp fragments to construct a paired-end (PE) library according to the Nextera XT sample preparation procedures (Illumina, San Diego, CA). We generated the PE reads of 150 bp using HiSeq X-Ten sequencer (Illumina, San Diego, California, USA).
In all, 3.53 Gb of raw sequence data were obtained. Reads were assembled into contigs using program CLC Genomics Workbench version 8.5.1 (CLC Inc, Arhus, Denmark). We annotated the complete plastome using the DOGMA pipeline (Wyman et al. Citation2004) and validated by comparing with the chloroplast genome of P. sinensis Sabine ex Lindl. (KU321892) (Liu et al. Citation2016). We determined transfer RNAs using tRNAscan-SE (Schattner et al. Citation2005). The annotated chloroplast genome of P. homogama was deposited into GenBank with the accession number MW167117.
The complete chloroplast genome of P. homogama was 154,677 bp in size with a typical quadripartite structure, containing a large single-copy (LSC) region of 85,299 bp and a small single-copy (SSC) region of 17816 bp. These two regions were separated by a pair of inverted repeat regions (IRs), each of 25,781 bp. A total of 130 functional genes were encoded, consisted of 86 protein-coding genes (PCG), 36 tRNA genes, and eight ribosomal RNA (rRNA) genes. Of these genes, 18 genes were duplicated in the IR region, including seven protein-coding genes, seven tRNA genes, and four rRNA genes. Eighteen genes contain one or two introns. The overall GC content of the chloroplast genome was 36.9%, whereas the corresponding values of the LSC, SSC, and IR regions were 34.8, 30.6, and 42.7%, respectively.
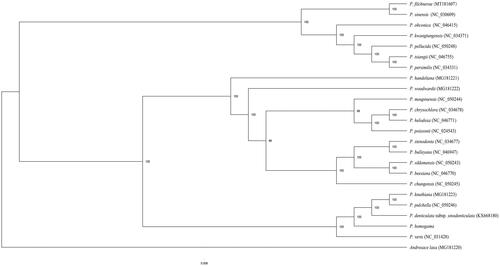
The maximum likelihood (ML) phylogenetic tree was reconstructed by RAxML (Stamatakis Citation2014) based on 23 complete chloroplast genomes of Primula species and Androsace laxa as outgroup. The phylogenetic results indicated that the sampled Primula species clustered as a monophyletic clade with high support (100%), and P. homogama formed a clade (100% bootstrap value) with P. knuthiana, P. pulchella, P. veris and P. denticulata subsp. sinodenticulata (). This published P. homogama chloroplast genome will provide useful information for phylogenetic and evolutionary studies in Primula.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statements
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW167117. The associated **BioProject**, **SRA**, and **Bio-Sample** numbers are SRX9518288, PRJNA678458, and SAMN16801853 respectively.
Additional information
Funding
References
- Doyle J. 1991. DNA protocols for plants: CTAB total DNA isolation. In: Hewitt GM, Johnston A, editor, Molecular techniques in taxonomy. Berlin: Springer-Verlag Press, p. 283–293.
- Ganders FR. 1979. The biology of heterostyly. New Zeal J Bot. 17(4):607–635.
- Hu CM, Kelso S. 1996. Primulaceae. In: Wu ZY, Raven PH, editors. Flora of China, Vol. 15. Beijing: Science Press; p. 240.
- Liu TJ, Zhang CY, Yan HF, Zhang L, Ge XJ, Hao G. 2016. Complete plastid genome sequence of Primula sinensis (Primulaceae): structure comparison, sequence variation and evidence for accD transfer to nucleus. PeerJ. 4:e2101.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and 825 snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server):W686–W689.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zhong L, Barrett SCH, Wang XJ, Wu ZK, Sun HY, Li DZ, Wang H, Zhou W. 2019. Phylogenomic analysis reveals multiple evolutionary origins of selfing from outcrossing in a lineage of heterostylous plants. New Phytol. 224(3):1290–1303.