Abstract
Camellia sinensis var. sinensis cultivar Tieguanyin (TGY) is an important Oolong tea variety in China. In this study, we reported a complete chloroplast (cp) genome based on the Illumina sequencing technology and combined de novo and reference-guided assembly strategies. The complete cp genome of ‘TGY’ displayed the regular quadripartite structure: a total of 157,126 bp in length, comprising a large single-copy (LSC, 86,904 bp) region, a small single-copy (SSC, 18,532 bp) region, and a pair of inverted repeats (IRs, 26,095 bp) regions. A lot of 132 predicted genes, including 87 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The overall GC content is 37.3%. Maximum likelihood (ML) phylogenetic tree involving 18 cp genomes of the Camellia genus revealed a relatively independent event of local domestication among three types of cultivars. The complete cp genome of ‘TGY’ provides an insight into tea plants for further understanding evolutionary research on tea plants.
Camellia sinensis var. sinensis cultivar Tieguanyin (‘TGY’, Chinese: ‘Iron Goddess of Mercy’), belonging to Camellia genus of the Theaceae, is an evergreen shrub originating from Anxi, Fujian province in nineteenth century. According to the historical records, ‘TGY’ was domesticated during Yongzheng Emperor in the Qing Dynasty (1723–1735 A.D.). In 1985, ‘TGY’ was certificated as an elite cultivar by National Crop Variety Approval Committee (NCVAC) in China with highly desirable traits such as strong resistance to cold and drought. As an important economic crop, it is widely cultivated in Fujian, Guangdong and Taiwan Province, which beverages made from leaves are most popular with excellent varieties and rich aroma in the oolong tea series (Lin et al. Citation2020). In 2016, TGY’s export volume reached 10,000 t, accounting for 50% of the total export of oolong tea in the international trade (Chen Citation2018). In addition to that, important secondary metabolites such as theanine and flavonoids are good for health benefits. ‘TGY’ is rich in caffeine, which is a good choice for relaxing while staying alert, during in times of stress. However, in recent years, some desirable traits in ‘TGY’ are obviously degraded, and the yield and quality also declined. Lots of genetic diversity was lost gradually due to clonal propagation and artificial selection. Thus, it is urgent to protect and improve this germplasm resource and make it become a good material for breeding improvement in cultivated tea plants. Recent studies have reported many tea plant nuclear genomes and organelle genomes (Huang et al. Citation2014; Xia et al. Citation2017, Citation2020; Wang et al. Citation2020; Zhang et al. Citation2020). However, ‘TGY’ due to high heterozygosity (2.8%), so far there are no suitable methods to resolve the genome. Therefore, it is still a gap to study genetic mechanisms and evolutionary history of ‘TGY’. Small chloroplast (cp) genome will be feasible in investigating the genetic mechanisms in C. sinensis. Here, we reported a complete cp genome sequence of C. sinensis var. sinensis cultivar TGY based on high-throughput sequencing technologies.
Young leaves of ‘TGY’ were collected from a single individual, planted in Anxi county located in Fujian Province, China (119.576708E, 27.215297N), and the specimens were preserved in the laboratory of the Haixia Institute of Science and Technology, Fujian Agriculture and Forestry University (specimen code: TGY20191011). The total genomic DNAs from the sample were isolated using DNeasy Plant Mini Kit (Qiagen, Germantown, MD) following the manufacturer’s instructions, and sequenced using the Illumina NovaSeq platform with 150-bp read length and 300–500 bp insert size. The complete cp genome of Camellia sinensis var. sinensis (GenBank database under the accession number: KJ806281), as a reference and the cp genome of ‘TGY’ was assembled by GetOrganelle pipeline (https://github.com/Kinggerm/GetOrganelle) (Jin et al. Citation2020). Total cp genome reads were extracted based on the close reference genome and conducted de novo assembly. The complete cp genome was annotated using Geseq (Tillich et al. Citation2017) and the circular genome map was visualized by OGDRAW (Greiner et al. Citation2019).
The complete cp genome of C. sinensis var. sinensis cultivar TGY (GenBank accession: MW148820) was 157,126 bp total in length, comprising of a large single-copy (LSC, 86,904 bp) region, a small single-copy (SSC, 18,532 bp) region, and a pair of inverted repeat (IR, each of size, 26,095 bp) regions. A total of 132 genes were annotated, including 87 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Total GC content was 37.3%.
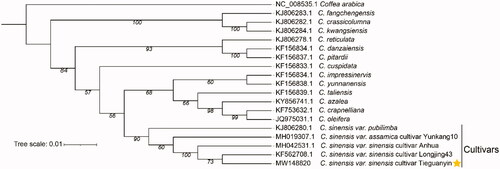
To investigate the evolutionary status of ‘TGY’ in the Camellia genus, 17 complete cp genomes of the Camellia genus and one outgroup taxa (Coffea arabica) were selected for phylogenetic analysis. All of them can be downloaded from NCBI GenBank. These genome sequences were multiply aligned using MAFFT (Katoh and Standley Citation2014). ML-based phylogenetic tree was constructed by RAxML (Stamatakis Citation2014) software with GTRCAT model and 1000 bootstrap replicates (). From the tree, we observed that the C. sinensis was obviously separated from these wild relative species, and the three domestic varieties (C. sinensis var. pubilimba, C. sinensis var. assamica, and C. sinensis var. sinensis) of tea plants clustered distinct branches, suggesting a relatively independent of local domestication. The complete cp genome of ‘TGY’ provides an insight into tea plants for further understanding evolutionary studies on C. sinensis and introduces a reference to facilitate crop improvement and enhances desirable traits in the ‘TGY’ breeding.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW148820. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA679706, SRR13090070, and SAMN16844543, respectively.
Additional information
Funding
References
- Chen MX. 2018. Development status and countermeasure of Tieguanyin industrialization in Anxi. Mod Agric Sci Technol. 15:262–264.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Huang H, Shi C, Liu Y, Mao S-Y, Gao L-Z. 2014. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evol Biol. 14:151.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2014. MAFFT: iterative refinement and additional methods. In: Russell DJ, editors. Multiple sequence alignment methods. Totowa (NJ): Humana Press; p. 131–146.
- Lin Y, Yu W, Zhou L, Fan X, Wang F, Wang P, Fang W, Cai C, Ye N. 2020. Genetic diversity of oolong tea (Camellia sinensis) germplasms based on the nanofluidic array of single-nucleotide polymorphism (SNP) markers. Tree Genet Genomes. 16:3.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang X, Feng H, Chang Y, Ma C, Wang L, Hao X, Li A, Cheng H, Wang L, Cui P, et al. 2020. Population sequencing enhances understanding of tea plant evolution. Nat Commun. 11(1):4447.
- Xia E, Tong W, Hou Y, An Y, Chen L, Wu Q, Liu Y, Yu J, Li F, Li R, et al. 2020. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into genome evolution and adaptation of tea plants. Mol Plant. 13(7):1013–1026.
- Xia E-H, Zhang H-B, Sheng J, Li K, Zhang Q-J, Kim C, Zhang Y, Liu Y, Zhu T, Li W, et al. 2017. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol Plant. 10(6):866–877.
- Zhang W, Zhang Y, Qiu H, Guo Y, Wan H, Zhang X, Scossa F, Alseekh S, Zhang Q, Wang P, et al. 2020. Genome assembly of wild tea tree DASZ reveals pedigree and selection history of tea varieties. Nat Commun. 11(1):3719.