Abstract
The complete mitochondrial genome of Dictyosoma burgeri collected from Yellow and Bohai Seas was determined by next-generation sequencing. The mitogenome is a circular molecule 16,513 bp in length, including the typical structure of 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and a control region. The TAS, central CSB, and CSB were detected in the control region. The gene contents of the mitogenome are identical to those observed in most bony fishes.
Dictyosoma burgeri (van der Hoeven, 1855) is mainly distributed along the coasts of China, Japan and Korea (Yatsu et al. Citation1978). It has been described in detail for the seasonal gonadal development and histological observation in Jeju, Korea (Jin et al. Citation2007). Hwang et al. investigated the function of plasma and cortisol in female during the annual reproductive cycle (Hwang et al. Citation2012). However, ribbed gunnel has not received much attention in population genetic studies. Next generation sequencing (NGS) has increased the speed and throughput capacities of DNA sequencing, thus dramatically reducing the overall cost of sequencing (Zhang et al. Citation2011). The appearance of NGS has foster the characterization of mitochondrial genomes in various species (Li et al. Citation2019), which can be used to further understand population genetics and evolutionary history of species (Zhang and Xian Citation2016). In the present study, we extracted DNA from muscle tissue of ribbed gunnel collected from Bohai and Yellow Seas in the September 2019 and conducted appropriate NGS analysis to isolate mt DNA sequences.
In this study, whole Genome Shotgun (WGS) strategy was adopted to construct library. Paired-end (PE) Sequencing was performed using Next-Generation Sequencing (NGS) based on Illumina MiSeq Sequencing platform. A5-miseq V20150522 (Coil et al. Citation2015) and SPAdesv3.9.0 (Bankevich et al. Citation2012) were used to assemble high-quality second-generation sequencing data from bottom to construct contig and scaffold sequences. The splice complete mitochondrial genome sequence was uploaded to the MITOS Web server (http://mitos.bioinf.uni-leipzig.de/) for functional annotation (Bernt et al. Citation2013). Genetic code selects the setting as 05-Inverterbrate, and the rest Settings follow the default parameters set by MITOS.
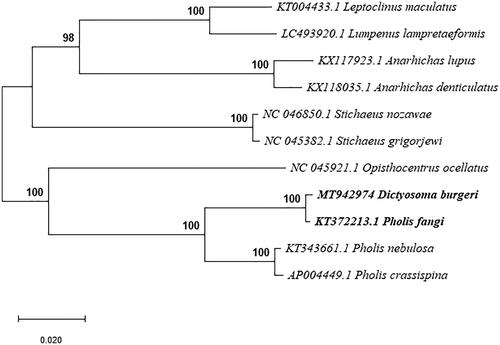
The complete mitogenome of Dictyosoma burgeri was 16,513 bp in length (Genbank accession no. MT942974), within the range of other teleost mitogenomes. As in other vertebrates (Miya et al. Citation2001), it contained 13 protein-coding genes, 2 rRNA genes (12S rRNA and 16S rRNA), 22 tRNA genes, and a control region. Like other bony fishes, most mitochondrial genes of D. burgeri were encoded on the H-strand, with only ND6 and eight tRNA (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, and Pro) genes encoded on the L-strand. Moreover, there were 3 overlapping reading frames in ND2 and ND5 genes, 4 in Cyt b gene. The ATPase 6 and ATPase 8 overlapped by 10 nucleotides, and ND4 and ND4L shared 7 nucleotides. ND5 and ND6 overlapped by 4 nucleotides on the opposite strand. ATG was the initiation codon of 12 out of the 13 protein coding genes (ND1, ND2, CO2, ATPase 8, ATPase 6, COIII, ND3, ND4L, ND4, ND5, ND6, and Cyt b), while the initiation codon of COI was GTG. TAA was the stop codon for 7 genes (ND2, COI, ATPase 8, ATPase 6, COIII, ND4L, and ND6), TAG was the stop codon for ND1, ND3 and ND5, the other genes had incomplete stop codons, either T (COII, ND4, and Cyt b), which were presumably completed as TAA by post-transcriptional polyadenylation (Ojala et al. Citation1981). The 12S and 16S ribosomal RNA genes of D. burgeri comprised 948 bp and 1668 bp, respectively. They were located between tRNAPhe and tRNALeu, and were separated by tRNAVal, as they were in other vertebrates (Gao et al. Citation2013). The 22 tRNA genes were interspersed in the genome and ranged in size from 66 to 74 bp and folded into cloverleaf secondary structures with normal base paring. The major noncoding region in D. burgeri was located between tRNAPro and tRNAPhe, and was determined to be 584 bp in length. The TAS, central CSB, and CSB were detected in the control region, which was similar to most bony fishes (Zhang et al. Citation2013). Phylogenetic analysis was accomplished by Mega X, using the complete sequence of D. burgeri and the other 10 closely related Blenniidae species. The Maximum Likelihood tree indicated that D. burgeri was sister species to Pholis fangi () and that the genus Pholis was not monophyletic.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/nuccore/MT942974. Associated BioProject, SRA, and BioSample accession numbers are https://www.ncbi.nlm.nih.gov/bioproject/PRJNA678842, https://www.ncbi.nlm.nih.gov/sra/ SAMN16815269, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589.
- Hwang IJ, Kim SY, Kim HB, Baek HJ. 2012. Changes in plasma sex steroid and cortisol levels during annual reproductive cycle of ribbed gunnel, Dictyosoma burgeri. Dev Reprod. 16(4):279–287.
- Gao T, Li N, Zhang Y, Shi P. 2013. The complete mitochondrial genome of Japanese sandeel Ammodytes personatus (Perciformes, Ammodytidae): rare structure in control region compared. Mitochondrial DNA. 24(4):320–322.
- Jin YS, Han JI, Park CB, Lee CH, Kim BH, Baek HJ, Kim HB, Lee YD. 2007. Reproductive cycle of ribbed gunnel, Dictyosoma burger. Kor J Ichthyol. 19:8–15.
- Li Q, Wang Q, Jin X, Chen Z, Xiong C, Li P, Zhao J, Huang W. 2019. The first complete mitochondrial genome from the family Hygrophoraceae (Hygrophorus russula) by next-generation sequencing and phylogenetic implications. Int J Biol Macromol. 122:1313–1320.
- Miya M, Kawaguchi A, Nishida M. 2001. Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol. 18(11):1993–2009.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290(5806):470–474.
- Yatsu A, Yasuda F, Taki Y. 1978. A new Stichaeid fish, Dictyosoma rubrimaculata from Japan, with notes on the geographic dimorphism in Dicyosoma burgeri. Jap J Ichthyol. 25(1): 40–50.
- Zhang H, Xian W. 2016. The complete mitochondrial genome of the larvae Japanese grenadier anchovy Coilia nasus (Clupeiformes, Engraulidae) from Yangtze estuary. Mitochondrial DNA Part A. 27(2):852–853.
- Zhang H, Zhang Y, Zhang X, Song N, Gao T. 2013. Special structure of mitochondrial DNA control region and phylogenetic relationship among individuals of the black rockfish, Sebastes schlegelii. Mitochondrial DNA. 24(2):151–157.
- Zhang J, Chiodini R, Badr A, Zhang G. 2011. The impact of next-generation sequencing on genomics. J Genet Genom. 38(3):95–109.