Abstract
The complete mitochondrial genome of Parastratiosphecomyia szechuanensis Lindner, 1954 was sequenced and analyzed in this study. The mitochondrial genome is 16,414 bp in length, including the 37 typical insect mitochondrial genes and a large control region. All PCGs end with complete termination codon TAA or TAG. Most PCGs initiated by standard start codon ATN, except for cox1 which starts with TCG. The phylogenetic analysis based on the nucleotide sequence data of 13 PCGs recovered the monophyly of Stratiomyidae and the sister relationship between Xylomyidae and Stratiomyidae.
The superfamily Stratiomyoidea belongs to the order Diptera, and is one of the key groups connecting Nematocera and Brachycera (Yang et al. Citation2014). Stratiomyidae is the larger of the two families of Stratiomyoidea with 282 genera and more than 30,000 species recorded in the world, has a relatively important evolutionary status (Zhou et al. Citation2017). However, there are only three mitochondrial genomes of Stratiomyidae in GenBank (as of 8 September 2020). Here, we sequenced the mitochondrial genome of Parastratiosphecomyia szechuanensis and investigated the phylogenetic relationships within Brachycera.
The adult specimens used in this study were collected from Luodian county (E106.7917, N25.4075), Guizhou province, China by Zaihua Yang in May 2020, and genetic material of this soldier fly (GZAF-2020-DS1435) was preserved in Insect Herbarium of Guizhou Academy of Forestry, Guiyang. The identification was based on morphological Characteristics (Yang et al. Citation2014). Next, the genomic DNA of P. szechuanensis was sequenced by next-generation sequencing (NGS). The complete mitochondrial genome was assembled and annotated using MitoZ (Meng et al. Citation2019). Maximum-likelihood phylogenetic analysis was performed by IQ-TREE version 1.6.10 (Vienna, Austria) (Nguyen et al. Citation2015) using the ultrafast bootstrap (UFB) approximation approach (Minh et al. Citation2013) with 8000 replicates.
The complete mitochondrial genome of P. szechuanensis (Genbank accession no. MW039153) is double-stranded circular DNA molecule with a size of 16,414 bp. The mitochondrial genome includes the 37 typical insect mitochondrial genes (13 protein-coding genes, 22 transfer RNA genes, and two ribosomal RNA genes) and a large AT-rich region (control region). The gene order of the newly sequenced Stratiomyidae is consistent with other known Stratiomyidae (Qi et al. Citation2017; Zhan et al. Citation2020). In the mitochondrial genome, the 23 genes (nine PCGs and 14 tRNAs) were encoded by the majority strand (J-strand) while 14 genes (four PCGs, eight tRNAs, and two rRNA genes) were encoded by the minority strand (N-strand). The overall nucleotide compositions of P. szechuanensis are A = 39%, C = 16.3%, G = 9.5%, and T = 35.2%, indicating the mitochondrial genome has a strong AT nucleotide bias. Most PCGs (atp6, atp8, cox2, cox3, cytb, nad1, nad2, nad3, nad4, nad4L, nad5, and nad6) initiated by standard start codon ATN (ATA/T/G/C), except for cox1 which starts with TCG. All PCGs end with complete termination codon TAA or TAG.
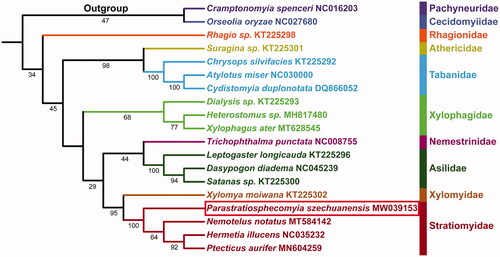
Phylogenetic analysis of 19 species of Diptera, including two outgroups from Nematocera, based on the concatenated dataset of 13 PCGs, yielded topology is similar to previously published phylogenies (Zhou et al. Citation2017; Ding and Yang Citation2020) (), but we have much larger taxon sample. The monophyly of all families (Tabanidae, Xylophagidae, Asilidae, and Stratiomyidae) represented by more than one species received support. The phylogenetic relationships among Brachycera is (Rhagionidae + ((Athericidae + Tabanidae) + (Xylophagidae + ((Asilidae + Nemestrinidae) + (Stratiomyidae + Xylomyidae))))). Furthermore, the sister relationship between Xylomyidae and Stratiomyidae is recovered with the high support value (BS = 95), in agreement with previously those found based on mitochondrial genome data (Zhou et al. Citation2017; Ding and Yang Citation2020).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, GenBank Accession Number: MW039153.
Additional information
Funding
References
- Ding SM, Yang D. 2020. Complete mitochondrial genome of Coenomyia ferruginea (Scopoli). (Diptera, Coenomyiidae). Mitochondrial DNA Part B. 5(3):2711–2712.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Qi YJ, Xu JY, Tian XX, Bai YC, Gu XS. 2017. The complete mitochondrial genome of Hermetia illucens (Diptera: Stratiomyidae). Mitochondrial DNA Part B. 2(1):189–190.
- Yang D, Zhang TT, Li Z. 2014. Stratiomyoidea of China. Beijing (China): Agricultural University Press; p. 1–870.
- Zhan QB, Zhan Y, Zhang SY, Peng XJ. 2020. The complete mitochondrial genome of the soldier fly Ptecticus aurifer. Mitochondrial DNA B Resour. 5(1):660–661.
- Zhou QX, Ding SM, Li X, Zhang TT, Yang D. 2017. Complete mitochondrial genome of Allognosta vagans (Diptera, Stratiomyidae). Mitochondrial DNA Part B. 2(2):461–462.