Abstract
Ceriagrion fallax is ubiquitous in south China and is particularly easy be found in some rice fields. In this study, we sequenced and analyzed the complete mitochondrial genome (mitogenome) of C. fallax. This mitogenome was 15,350 bp long and encoded 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs) and two ribosomal RNA unit genes (rRNAs). The nucleotide composition of the mitogenome was biased toward A and T, with 74.0% of A + T content (A 42.1%, T 31.9%, C 14.6%, G 11.4%). Gene order was conserved and identical to most other previously sequenced Zygoptera dragonflies. Most PCGs of C. fallax have the conventional start codons ATN (seven ATG, two ATT, and two ATC), with the exception of nad3 and nad1 (TTG). Except for four PCGs (cox1, cox2, cox3, and nad5) end with the incomplete stop codon T––, all other PCGs terminated with the stop codon TAA. Phylogenetic analysis showed that C. fallax got together with the same family species (Agriocnemis femina, Enallagma cyathigerum, Ischnura elegans, Ischnura pumilio) with high support value. The relationships (Megapodagrionidae + ((Calopterygidae + (Euphaeidae + Pseudolestidae)) + (Coenagrionidae + Platycnemididae))) were supported within Zygoptera.
The insect family Coenagrionidae is placed in the suborder Zygoptera which is known as damselflies. Coenagrionidae is a diverse and ancient group, with the first fossils assignable to this family dated to the early Cretaceous (Chippindale et al. Citation1999). More than 1100 species are in this family, including six subfamilies: Agriocnemidinae, Argiinae, Coenagrioninae, Ischnurinae, Leptobasinae, and Pseudagrioninae. Ceriagrion fallax Ris 1914, one of species within Coenagrionidae, is common in South China. Females and males of this species have different external morphology characters. Here, we sequenced and analyzed the complete mitochondrial genome of C. fallax.
Specimens of C. fallax were collected from Yongxin County, Jiangxi Province, China (26°54′N, 114°14′E, July 2019) and were stored in Entomological Museum of Qinghai Academy of Agriculture and Forestry Sciences (Accession no. QHAF-CF03). Total genomic DNA was extracted from tissues using DNeasy DNA Extraction kit (Qiagen, Hilden, Germany). A pair-end sequence library was constructed and sequenced using Illumina HiSeq 2500 platform (Illumina, San Diego, CA), with 150 bp pair-end sequencing method. A total of 20.1 million reads were generated and had been deposited in the NCBI Sequence Read Archive (SRA) with accession number SRR12807218. With the mitochondrial genome of Pseudolestes mirabilis (FJ606784) employed as reference, raw reads were assembled using MITObim v 1.7 (Hahn et al. Citation2013). By comparison with the homologous sequences of other Zygoptera species from GenBank, the mitogenome of C. fallax was annotated using software GENEIOUS R11 (Biomatters Ltd., Auckland, New Zealand).
The complete mitogenome of C. fallax is 15,350 bp in length (GenBank accession no. MW092110), and containing the typical set of 13 protein-coding, two rRNA and 22 tRNA genes, and one non-coding AT-rich region. The nucleotide composition of the mitogenome was biased toward A and T, with 74.0% of A + T content (A 42.1%, T 31.9%, C 14.6%, and G 11.4%). Gene order was conserved and identical to most other previously sequenced Zygoptera dragonflies (Lin et al. Citation2010; Lorenzo-Carballa et al. Citation2014; Zhang et al. Citation2017; Lan et al. Citation2019; Song et al. Citation2019). Two rRNA genes (rrnL and rrnS) locate at trnL1/trnV and trnV/control region, respectively. The lengths of rrnL and rrnS in C. fallax are 1300 and 752 bp, with the AT contents of 77.4% and 75.1%, respectively. All 22 tRNA genes were predicated in this study and vary from 64 bp (trnC) to 72 bp (trnK). Most PCGs of C. fallax have the conventional start codons ATN (seven ATG, two ATT, and two ATC), with the exception of nad3 and nad1 (TTG). Except for four PCGs (cox1, cox2, cox3, and nad5) end with the incomplete stop codon T−, all other PCGs terminated with the stop codon TAA.
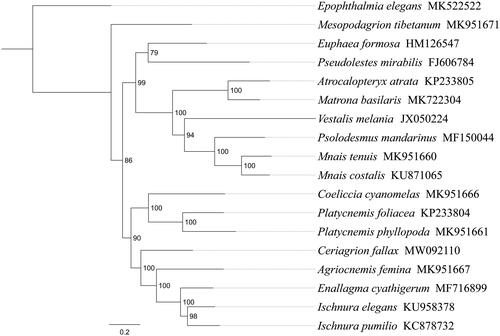
Phylogenetic analysis was performed based on the nucleotide sequences of 13 PCGs from 18 Odonata species. Phylogenetic tree was constructed through raxmlGUI 1.5 (Silvestro and Michalak Citation2012). Results showed that 17 Zygoptera species divided into three main clades, the new sequenced species C. fallax got together with the same family species (Agriocnemis femina, Enallagma cyathigerum, Ischnura elegans, Ischnura pumilio) with high support value (), indicating the monophyly of Coenagrionidae could be confirmed. Within Zygoptera, the relationships (Megapodagrionidae + ((Calopterygidae + (Euphaeidae + Pseudolestidae)) + (Coenagrionidae + Platycnemididae))) were supported. In conclusion, the mitogenome of C. fallax is sequenced in this study and can provide essential and important DNA molecular data for further phylogenetic and evolutionary analysis of Coenagrionidae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI (National Center for Biotechnology Information) at https://www.ncbi.nlm.nih.gov/, reference number MW092110, SRR12807218.
Additional information
Funding
References
- Chippindale PT, Dave VK, Whitmore DH, Robinson JV. 1999. Phylogenetic relationships of North American damselflies of the genus Ischnura (Odonata: Zygoptera: Coenagrionidae) based on sequences of three mitochondrial genes. Mol Phylogenet Evol. 11(1):110–121.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):*e129–e129.
- Lan DY, Shen SQ, Cai YY, Wang J, Zhang JY, Storey KB, Yu DN. 2019. The characteristics and phylogenetic relationship of two complete mitochondrial genomes of Matrona basilaris (Odonata: Zygoptera: Calopterygidae). Mitochondr DNA B. 4(1):1745–1747.
- Lin CP, Chen MY, Huang JP. 2010. The complete mitochondrial genome and phylogenomics of a damselfly, Euphaea formosa support a basal Odonata within the Pterygota. Gene. 468(1–2):20–29.
- Lorenzo-Carballa MO, Thompson DJ, Cordero-Rivera A, Watts PC. 2014. Next generation sequencing yields the complete mitochondrial genome of the scarce blue-tailed damselfly, Ischnura pumilio. Mitochondrial DNA. 25(4):247–248.
- Silvestro D, Michalak I. 2012. RaxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12(4):335–337.
- Song N, Li X, Yin X, Li X, Yin J, Pan P. 2019. The mitochondrial genomes of palaeopteran insects and insights into the early insect relationships. Sci Rep. 9(1):1–11.
- Zhang L, Wang XT, Wen CL, Wang MY, Yang XZ, Yuan ML. 2017. The complete mitochondrial genome of Enallagma cyathigerum (Odonata: Coenagrionidae) and phylogenetic analysis. Mitochondr DNA B. 2(2):640–641.