Abstract
Paramesotriton aurantius (Caudata: Salamandridae) is a new species that found in southeastern China. Its complete mitochondrial genome (mitogenome) sequence was 16,313 bp in length with with A + T contents of 60.9%, and contained 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, one control region (D-loop), and one non-coding region. Our molecular tree showed that P. aurantius was positioned near P. hongkongensis, and formed a clade with other Paramesotriton species. The first complete mitogenome sequence of P. aurantius could provided fundamental data for resolving phylogenetic and genetic problems related to genus Paramesotriton.
There are 14 species in the genus of Paramesotriton in the world, and they are distributed in Northern Vietnam and southwest-central and southern China (Frost Citation2020). The main adult external morphological traits of Paramesotriton species are skin rough, dorsal warts small and scattered evenly wart on the sides of body relatively large and distinct (Fei and Ye Citation2016). This genus consists of two species groups (P. caudopunctatus group and P. chinensis group) (Yuan et al. Citation2014), but the phylogenetic relationship between the two groups has not been well solved. In 2016, Paramesotriton aurantius (Yuan et al. Citation2016) is a new species of the genus Paramesotriton that found in southeastern China (Yuan et al. Citation2016). Until now, four geographical populations of this species were known (Yuan et al. Citation2016; Liu et al. Citation2019). The mitochondrial genome (mitogenome) as the molecular marker could utilized to examine the phylogeny of Paramesotriton species. In Genbank, three complete mitogenome of Paramesotriton species are available (Zhang et al. Citation2005; Yang et al. Citation2017; Zhang et al. Citation2018), but not containing P. aurantius. In this study, we determined the complete mitogenomes via next-generation sequencing, and compared the sequence with those of other species of Salamandridae to analyze the phylogenetic relationship among them.
The specimen of P. aurantius (specimen voucher: LSU20200810WDY01) was captured from Wangdongyang (27.702646°N 119.626208°E; elevation: 1045 m), Lishui City, Zhejiang Province, China, and stored at the Museum of Laboratory of Amphibian Diversity Investigation at Lishui University by 75% alcohol in specimen bottle. Genomic DNA extraction and next-generation sequencing were described in previous publication (Chen et al. Citation2020). The mitogenomes were sequencing by Illumina NovaSeq 6000 platform. Raw data (22.09 G) was deposited in the NCBI’s Sequence Read Archive database, and clean data (21.35 G) subjected to de novo assembled by the NOVO Plasty 3.7 (Dierckxsens et al. Citation2017) to produce a closed circular form of complete mitogenome.
The complete mitogenome sequence of P. aurantius is 16,313 bp in length with with A + T contents of 60.9% (32.9% A, 28.0 T%, 14.9% G, and 24.2% C). The mitogenome was annotated by MITOS WebServer (http://mitos2.bioinf.uni-leipzig.de/) (Matthias et al. Citation2013) and tRNAscan-SE version 2.0 (http://trna.ucsc.edu/tRNAscan-SE/) (Lowe and Chan Citation2016), and included 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA (12S rRNA and 16S rRNA) genes, one control region (D-loop), and one non-coding region. Twenty-eight out of 37 genes were encoded on the H-strand, while 9 were located on the L-strand. The 12S rRNA (929 bp) and 16S rRNA genes (1,559 bp) were located between tRNAPhe and tRNAVal, and between tRNAVal and tRNALeu, respectively. Except COI gene which was began with GTG, other 12 PCGs were initiated with ATG as start codon. The stop codon of the PCGs was TAA (ND2, ND3, ND4L, ND5, ATP6, ATP8, and COI), TAG (ND1), AGA (ND6), TA (ND4 and CYTB), and T (COII and COIII). The complete mitogenome of P. aurantius had the same gene arrangement and similar compositions according with other Paramesotriton species (Zhang et al. Citation2005; Yang et al. Citation2017; Zhang et al. Citation2018).
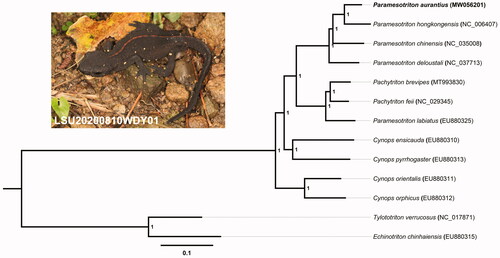
We used MrBayes v3.2.2 software to construct a Bayesian tree (with 1,000,000 generations, sampling every 1000 generations and discarded 1000 trees as burn-in) containing complete mitogenomes of 13 species to further explore taxonomic status of P. aurantius and the phylogenetic relationship within Paramesotriton. The phylogenetic tree showed that P. aurantius is sister to P. Hongkongensis and that the Paramesotriton genus is monophyletic (). This result was supported by previous phylogenetic studies (Yuan et al. Citation2014, Citation2016). The first complete mitogenome sequence of P. aurantius from our study will contribute to the further understanding of the genus Paramesotriton.
Figure 1. The Bayesian tree base on 13 concatenated mitochondrial PCGs of 13 species from the family Salamandridae. Echinotriton chinhaiensis and Tylototriton verrucosus were chosen as the outgroups. Numbers at the nodes represent Bayesian posterior probabilities. Samples sequenced in the present study are highlighted in bold.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at: https://www.ncbi.nlm.nih.gov/nuccore/MW056201. SRA and BioSample accession numbers are https://www.ncbi.nlm.nih.gov/sra/SRR12750572, https://www.ncbi.nlm.nih.gov/biosample/SAMN16326665, respectively.
Additional information
Funding
References
- Chen QR, Lin YF, Ma L, Ding GH, Lin ZH. 2020. Partial mitochondrial genome of the Sanchiang Tree Toad Hyla sanchiangensis (Anura: Hylidae). Mitochondrial DNA B. 5(3):2682–2683.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Fei L, Ye CY. 2016. Amphibians of China Volume (I). Beijing: Science Press; p. 349–354.
- Frost DR. 2020. Amphibian species of the world: an online reference, version 6.0. New York (NY): American Museum of Natural History. [accessed 2020 October 14]. http://research.amnh.org/herpetology/amphibia/index.php/.
- Liu RL, Zhou JJ, Liu KC, Jin W, Ye WJ, Xu XX. 2019. Paramesotriton aurantius (Caudata: Salamandridae) Found in Zhejiang Province. Chin J Zool. 54:117–122.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Matthias B, Alexander D, Frank J, Fabian E, Catherine F, Guido F, Joern P, Martin M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Yang J, Yu LP, Zhang SY, Liu RL, Chen CS, Lin HD. 2017. The complete mitochondrial genome of Paramesotriton chinensis (Caudata: Salamandridae) and phylogenetic studies of Paramesotriton. Mitochondrial DNA B. 2(1):289–290.
- Yuan ZY, Zhao HP, Jiang K, Hou M, He L, Murphy RW, Che J. 2014. Phylogenetic relationships of the genus Paramesotriton (Caudata: Salamandridae) with the description of a new species from Qixiling Nature Reserve, Jiangxi, southeastern China and a key to the species. Asian Herpetol Res. 5:67–79.
- Yuan ZY, Wu YK, Zhou JJ, Che J. 2016. A new species of the genus Paramesotriton (Caudata: Salamandridae) from Fujian, southeastern China. Zootaxa. 4205(6):zootaxa.4205.6.3–563.
- Zhang P, Zhou H, Chen YQ, Liu YF, Qu LH. 2005. Mitogenomic perspectives on the origin and phylogeny of living amphibians. Syst Biol. 54(3):391–400.
- Zhang MW, Han FY, Ye J, Ni QY, Li Y, Yao YF, Xu HL. 2018. The entire mitochondrial genome of Vietnam Warty Newt Paramesotriton deloustali (Salamandridae: Paramesotriton) with a new distribution record from China. Conservation Genet Resour. 10(3):551–554.
