Abstract
The complete chloroplast (cp) genome of Neopallasia pectinata was sequenced and analyzed in this study. It was 150,766 bp in length and has a typical circular structure, including a large single copy (LSC) with 82,605 bp, two inverted repeats (IRs) with 24,944 bp, and a small single copy (SSC) with 18,273 bp. The phylogenetic analysis of N. pectinata and its related taxa was conducted depended on the complete cp-genome sequences. The maximum likelihood tree indicates a close relationship between Chrysanthemum and Neopallasia. The cp-genome of N. pectinata is useful for future phylogenetic studies of Asteraceae.
As a monotypic genus of the family Asteraceae, Neopallasia pectinata (Pall.) Poljakov is mainly distributed in China, Kazakhstan, Mongolia, and Russia (Shi et al. Citation2010). According to the micromorphological characteristics of the seeds, Neopallasia should be considered as an independent genus (Zhong et al. Citation2006). However, the phylogenetic analysis by ITS and EST showed that it was closely related to Artemisia and considered that N. pectinata should be included in Artemisia (Pellicer et al. Citation2011). There is no study about the cp-genome of N. pectinata. The complete chloroplast genome of N. pectinata was firstly reported and described in this study. It would provide genomic resources for further study on the phylogenetic relationship of Asteraceae plants.
The plant material (leaves of the mature plant) was collected at Tongde county in Qinghai Province (Geographic coordinates 35°29′58″N, 100°09′31″E; Altitude 2700 m). The voucher specimen (zhang2014073) was deposited in Qinghai-Tibetan Plateau Museum of Biology (HNWP), Northwest Institute of Plateau Biology, Chinese Academy of Sciences. The total genomic DNA of N. pectinata was extracted from the dry leaf with the modified CTAB method (Doyle Citation1987). The complete cp-genome of N. pectinata was sequenced by Illumina NovaSeq 6000 (Illumina Inc., San Diego, CA, USA) using 150 bp paired-end sequencing. 4.96 Gb sequencing data that contained 60,915,600 bp for paired-end reads were generated. The cp-genome was assembled with Unicycler v0.4.8 (Wick et al. Citation2017), and assembly script from Github (Wang et al. Citation2018). The final cp-genome of N. pectinata was annotated using the GeSeq annotation tool (Michael et al. Citation2017) with the default parameters, whereas the transfer RNA genes were further verified by tRNAscan-SE v.2.0.3 (Lowe and Chan Citation2016), and the result was corrected manually. The final cp-genome of N. pectinata was submitted to the GenBank (Accession Number: MW007388).
The cp-genome of N. pectinata has a typical circular structure with 150,766 bp, including a large single copy (LSC) with 82,605 bp, two inverted repeats (IRs) with 24,944 bp, and a small single copy (SSC) with 18,273 bp. The overall GC content is 37.48%. It contains 113 genes, including 80 unique protein-coding genes, 4 unique rRNA genes, and 29 unique tRNA genes. 18 genes were duplicated in the IR region, including 4 rRNA genes, 7 tRNA genes, and 7 protein-coding genes.
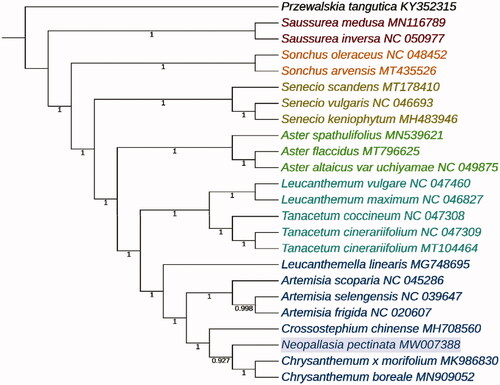
The nucleotide sequences of 79 protein-coding sequences (CDS) of 24 species were used for phylogenetic analysis. In addition to the cp-genome of N. pectinata, 23 other species were downloaded from NCBI, including 22 species in 10 genera of family Asteraceae and one species of family Przewalskia as the outgroup. The Bayesian inference tree was reconstructed by MrBayes v3.2.7 (Ronquist and Huelsenbeck Citation2003), with two independent runs each with four Markov chains, 1,000,000 generations starting from a random tree, and tree sampling of every 1000 generations. The result indicated that there is a close relationship between Chrysanthemum, Crossostephium, and Neopallasia (). Our study supports that Neopallasia is a monotypic genus of the family Asteraceae. And the complete cp-genome sequence of N. pectinata will provide a useful resource for future phylogenetic studies of Asteraceae.
Disclosure statement
The authors declare no conflicts of interest and are responsible for the content.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession NO. MW007388. The associated BioProject, Bio-Sample, and SRA number are PRJNA687065 and SAMN17132339, and PRJNA687065 respectively.
Additional information
Funding
References
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Michael T, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Pellicer J, Valles J, Korobkov AA, Garnatje T. 2011. Phylogenetic relationships of Artemisia subg. Dracunculus (Asteraceae) based on ribosomal and chloroplast DNA sequences. Taxon. 60(3):691–704.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Shi Z, Chen YL, Chen YS, Lin YR, Liu SW, Ge XJ, Gao TG, Zhu SX, Liu Y, Yang QE, et al. 2010. Flora of China. Vol. 20–21. Beijing: Science Press; p. 748.
- Wang Y, Feng S, Li S, Tang D, Chen Y, Chen Y, Zhou B. 2018. Assembly of chloroplast genomes with long- and short-read data: a comparison of approaches using Eucalyptus pauciflora as a test case. BMC Genomics. 19(1):15.
- Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. Plos Comput Biol. 13:22.
- Zhong SH, Li HX, Sheng CY. 2006. Micromorphological features of seed from six genera plants in tribe anthemideae. Acta Bot Boreali-Occidentalia Sin. 26:2214–2219.