Abstract
The root of Bupleurum marginatum var. stenophyllum (H. Wolff) Shan & Yin Li (Apiaceae), a new substitution for the popular Chinese medicinal material, Bupleuri Radix (Chai hu), is not easily distinguishable via traditional methods. The complete chloroplast genome sequence of B. marginatum var. stenophyllum was characterized using next-generation sequencing and the de novo assembly method. The complete genome was 155,576 bp in length and contained two inverted repeat (IR) regions of 26,311 bp, a large single-copy (LSC) region of 85,351 bp, and a small single-copy (SSC) region of 17,603 bp. It encoded 113 unique genes consisting of 79 protein-coding genes (PCGs), 30 transfer RNA genes, and four ribosomal RNA genes. Importantly, three genes (petB, petD and rps16) with small exon, and one trans-splicing gene (rps12) were correctly annotated. The overall GC content of the B. marginatum var. stenophyllum chloroplast genome is 37.7%. The phylogenetic analyses indicated that B. marginatum var. stenophyllum was closely related to B. marginatum. Moreover, many genetic information sites were available for distinguishing B. marginatum var. stenophyllum from the official ‘Chai hu’ plant sources, B. scorzonerifolium Willd. and B. chinense DC.
Bupleurum marginatum var. stenophyllum (H. Wolff) Shan & Yin Li is a perennial plant belonging to Bupleurum L. of the family Apiaceae (Wu et al. Citation2005). The roots of this species are often referred to as ‘Zang chai hu’ in the literature and frequently used in folk medicine for the treatment of various diseases, such as cough, fever, and influenza in Tibet, as well as the Yunnan, Guizhou, and Gansu provinces of China (Fang et al. Citation2017; Guo et al. Citation2018). Considering the popularity of Bupleuri Radix (Chai hu) in the current medicinal market, as well as the stable supply and substantial yield, ‘Zang chai hu’ represents a commercial source for regional substitutes of ‘Chai hu’ or other local medicinal purposes (Guo et al. Citation2018). Even though the resources, phytochemistry, pharmacology, and related research associated with this medicine are increasing (Ding et al. Citation2016; Fang et al. Citation2017; Yuan et al. Citation2017), no comprehensive genomic analysis currently exists. Therefore, the chloroplast genome of B. marginatum var. stenophyllum and the analysis of its genetic information and phylogenetic relationships within the Apiaceae family are explored here to improve the development and utilization of this resource.
Fresh B. marginatum var. stenophyllum leaf materials were collected from Lintao County, Gansu Province, China (N35°14′5.67″, E103°44′1.44″), and allocated the ID, GSC03. A voucher specimen was deposited in the herbarium of the Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, with an assigned voucher number of HPCH0002. The code of this herbarium was denoted by ‘IMD’ (NYBG: https://www.nybg.org/). A CTAB-based extraction method, modified according to a technique delineated by Porebski et al. (Porebski et al. Citation1997), was employed to extract total genomic DNA from the fresh leaf samples. A NanoDrop 2000 ultra-micro spectrophotometer was used to measure the quality of the DNA (Thermo Fisher Scientific Inc., USA), and next quantified using Qubit 4.0 (Thermo Fisher Scientific Inc., USA). This process was followed by creating ∼350 bp PCR-free libraries from the extracted genomic DNA according to the TruSeq DNA PCR-free library preparation guide. Sequencing was performed using the Illumina NovaSeq platform, generating 5,669,517 paired-end reads, totaling 1.7 Gb. Trimmomatic v0.38 (Bolger et al. Citation2014) was employed to filter the low-quality reads and the sequencing adapter. A NOVOPlasty toolkit (Dierckxsens et al. Citation2016) was used to compile the complete B. marginatum var. stenophyllum chloroplast genome after which the CPGAVAS2 webserver was employed for separate annotation (Shi et al. Citation2019).
The complete chloroplast genome of B. marginatum var. stenophyllum displayed a total sequence length of 155,576 bp (GenBank accession no. MT075712) and a GC content of 37.7%, which was similar to that of other Bupleurum genus species, such as B. chinense DC (Zhang et al. Citation2019). Furthermore, it featured the typical quadripartite structure of most angiosperm chloroplasts, containing two IR regions of 26,311 bp, an LSC region of 85,351 bp, and an SSC region of 17,603 bp. Finally, a total of 113 unique genes were successfully annotated in the complete chloroplast genome of B. marginatum var. stenophyllum, comprising 79 PCGs, 30 transfer RNA genes, and four ribosomal RNA genes. 17 unique genes (11 PCGs and six transfer RNA genes) contain introns. Three genes (petB, petD, and rps16) have small exon, and one gene (rps12) has been recognized as a trans-splicing gene. Codon usage analysis showed that 64 types of codons were used for the 79 PCGs and the AAA codon encoding isoleucine was the most frequently used.
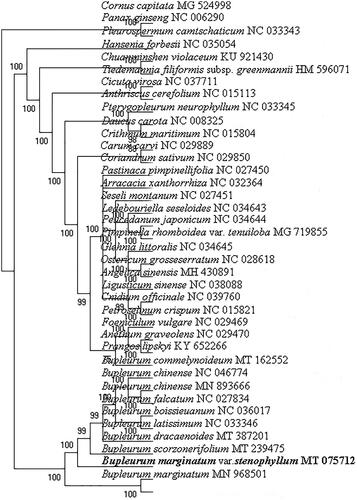
The phylogenetic position of B. marginatum var. stenophyllum was confirmed using RAxML v8.0.0 (Stamatakis Citation2014) to create a maximum-likelihood tree (with 1000 bootstrap replicates) using the GTR + I + G model based on its and other 37 chloroplast genomes (). The B. marginatum var. stenophyllum was closely clustered with B. marginatum and significantly separated from other species, including the official plant sources of ‘Chai hu,’ B. scorzonerifolium Willd. and B. chinense DC. The phylogenetic analysis results were consistent with the morphological observations, indicating that B. marginatum var. stenophyllum represented a separate species (Shan and Li Citation1974). Furthermore, the recent comparative analysis of chemical constituents in B. chinense and B. marginatum var. stenophyllum demonstrated that the compounds in B. marginatum var. stenophyllum differed from those in B. chinense (Yang et al. Citation2019). Therefore, substituting B. marginatum var. stenophyllum is a potential risk for using Chinese patent medicines containing ‘Chai hu’ (Liu et al. Citation2019). In conclusion, the complete chloroplast genome of B. marginatum var. stenophyllum provides essential DNA data for distinguishing B. marginatum var. stenophyllum from B. scorzonerifolium, and B. chinense.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT075712. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA682316, SRR13195839, and SAMN16990902, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18–e18.
- Ding C, Xu Y, Ma XX, Wang ZP, Mao WW, Zhang LL. 2016. Identification on five kinds of easily confused Bupleurum medicinal materials. J Chin Med Mat. 39(9):1975–1981.
- Fang W, Yang YJ, Guo BL, Cen S. 2017. Anti-influenza triterpenoid saponins (saikosaponins) from the roots of Bupleurum marginatum var. stenophyllum. Bioorg Med Chem Lett. 27(8):1654–1659.
- Guo JQ, Yang YJ, Fang W, Guo BL. 2018. Saikosaponins content in Bupleurum marginatum var. stenophyllum and cultivated B. chinese and B. marginatum. Mod Chin Med. 20(1):34–38.
- Liu XX, Chen F, Lin JF, Qiao L, Huang GK, et al. 2019. Discussion on quality control methods for chinese patent medicines containing Bupleurum. Chin Pharma J. 54:1452–1456.
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15(1):8–15.
- Shan RH, Li Y. 1974. On the Chinese species of Bupleurum L. Acta Phytotaxonomica Sinica. 12(3):262–294.
- Shi LC, Chen HM, Jiang M, Wang LQ, Wu X, Huang LF, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wu Z, Raven P, Hong D. 2005. Flora of China. Volume 14: Apiaceae through Ericaceae. Beijing: Science Press.
- Yang YJ, Zheng W, Guo JQ, Fang W, Ma BP, et al. 2019. Comparative analysis on chemical constituents in Bupleurum chinense, B. marginatum, B. marginatum var. stenophyllum and B. smithii var. parvifolium. China J Chin Materia Medica. 44(2):332–337.
- Yuan BC, Li WD, Ma YS, Zhou S, Zhu LF, Lin RC, Liu Y. 2017. The molecular identification of Bupleurum medicinal species and the quality investigation of Bupleuri Radix. Yao Xue Xue Bao. 52(1):162–171.
- Zhang F, Zhao ZY, Yuan QJ, Chen SQ, Huang LQ. 2019. The complete chloroplast genome sequence of Bupleurum chinense DC (Apiaceae). Mitochondrial DNA B Resour. 4(2):3665–3666.