Abstract
Bupleurum hamiltonii has a thin, wood, grayish-yellow root and is reputed to possess medicinal value. This study employs the de novo method using high-throughput sequencing data to assemble the complete chloroplast genome for this species. The total B. hamiltonii genome is 155,320 bp in length, containing a large single-copy region of 85,280 bp, a small single-copy region of 17,468 bp, a pair of inverted repeat regions of 26,286 bp, and has a GC content of 37.8%. The genome encodes 113 unique genes, of which there are 79 protein-coding, 30 tRNA, and four rRNA genes. In addition, 18 genes contained introns, petB, petD, and rpl16 are coded for by two exons, and rps12 is identified as a trans-splicing gene. Phylogenetic analysis results strongly suggest that B. hamiltonii is closely related to B. marginatum. This study provides valuable genetic information to facilitate reliable identification.
Bupleurum L. is a large genus in the family, Apiaceae, with about 180 species widely distributed in the North Temperate Zone (She and Watson Citation2005). In China, 44 species, 17 varieties, and seven forma of this genus have been reported, of which 25 species, eight varieties, and three forma have been recorded as resources for the traditional Chinese herbal medicine Radix Bupleuri (‘Chai hu’) (Pan et al. Citation2002). However, the official original plants listed for ‘Chai hu’ in Chinese Pharmacopeia only include B. chinense DC and B. scorzonerifolium Willd. Given the high species diversity and reputable medicinal value of this genus, the taxonomy assignment, chemical profiling, and biological activity of many species in the genus, Bupleurum have received considerable attention (Wang et al. Citation2011b). Several easily confused species are used as folk medicine in some local areas (Liu Citation2008; Yang et al. 2007). As an annual or short-lived perennial plant with a thin root and characteristic fruit (She and Watson Citation2005), Bupleurum hamiltonii N. P. Balakrishnan is commonly documented as ‘Xiao chai hu’ (Liu Citation2008; Ma et al. Citation2010; Wang et al. Citation2011a), which constitutes a similar name as the famous composite formula ‘xiao chai hu tang.’ In addition, B. chinense, widely cultivated in Gansu, is also commonly called ‘Xiao chai hu’ (Zhang Citation2020), which may lead to the misuse of B. hamiltonii and B. chinense.
Fresh B. hamiltonii leaves were collected from Supu Town, Qianxi County, Bijie City, Guizhou Province (N26°59′39.75″, E106°21′29.02″). The voucher specimen with the number of HPCH0003 was placed in the herbarium at the Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College (IMD). Total genomic DNA was isolated from the leaves using modified CTAB-based extraction (Porebski et al. Citation1997). The quantity and quality of the DNA were examined using the Qubit 4.0 (Thermo Fisher Scientific Inc., USA). After the total DNA was sheared, the TruSeq DNA PCR-free library preparation guide was used to generate a library with an average insert size of 270 bp. The Illumina NovaSeq platform was employed to conduct high-throughput sequencing, and approximately 3.4 GB of raw data were generated with 150 bp paired-end read lengths. Low-quality reads and the sequencing adapter were filtered using Trimmonmatic v0.38 (Bolger et al. Citation2014), and only the ‘paired’ output files where both reads survived processing were used for the subsequent analysis. The de novo assembly of the complete chloroplast genome was completed using the NOVOPlasty toolkit (Dierckxsens et al. Citation2016), and the average genome sequencing depth was 516. The genome annotation for protein-coding, rRNA, and tRNA genes was performed with CPGAVAS2 (Shi et al. Citation2019) on the website www.herbalgenomics.org/cpgavas2. The gene and repeat element map of the annotated B. hamiltonii chloroplast genome was generated synchronously during the annotation process.
The chloroplast genome sequence of B. hamiltonii is 155,320 bp in length and shows a typical quadripartite structure like most angiosperm plants, with two reverse repeated regions (IRa and IRb) of 26,286 bp in length. The repeat regions divide the entire chromosome into two single-copy regions, namely a small single-copy region (SSC) and a large single-copy region (LSC) with 17,468 bp and 85,280 bp, respectively (GenBank accession no. MW262986). The total GC content of the chloroplast genome was 37.8% and encodes 113 unique genes. Of these, 79 were protein-coding, 30 represented tRNA, and four are rRNA genes. Furthermore, 18 genes contain introns, 15 (nine protein-coding and six tRNA genes) contain one intron and three (rps12, ycf3, and clpP) two introns. Moreover, petB, petD, and rpl16 are coded by two exons, while rps12 is identified as a trans-splicing gene.
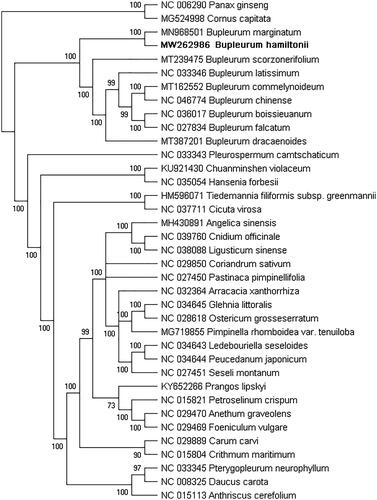
To confirm the phylogenetic location of B. hamiltonii in the genus Bupleurum of the Apiaceae, a total of 37 complete chloroplast genomes were used for the phylogenetic analysis using the Maximum Likelihood (ML) method with RAxML v8.0.0 (Stamatakis Citation2014) and 1000 bootstrap replicates. Panax ginseng of the family, Araliaceae, and Cornus capitata of the family, Cornaceae, were designated as the outgroups. Based on the results of the phylogenetic analysis, the nine species of the genus Bupleurum were fully supported in a monophyletic clade with a bootstrap value of 100 (), which was consistent with previous studies (Yang et al. Citation2007). The ML tree showed that B. hamiltonii and B. marginatum were resolved in the same clade, while B. chinense, B. scorzonerifolium, and five additional species of the genus Bupleurum were sister in position. The complete chloroplast genome of B. hamiltonii will provide valuable genetic information for further study involving genetic diversity, especially regarding reliable identification.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Chloroplast data supporting this study are openly available in GenBank at nucleotide database, https://www.ncbi.nlm.nih.gov/nuccore/ MW262986, Associated BioProject, https://www.ncbi.nlm.nih.gov/bioproject/ PRJNA682316, BioSample accession number at https://www.ncbi.nlm.nih.gov/biosample/ SAMN16987561 and Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra/ SRR13189612.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18–e18.
- Liu L. 2008. Oligonucleotide DNA chip based on ITS region polymorphism for identification of Bupleurum plants-derived crude drugs. Master’s Thesis. Southern Medical University, Guanzhou.
- Ma XQ, Wang CB, He XJ. 2010. Micromorphological features of pericarp surface of Bupleurum L.(Apiaceae) in China and its taxonomic significance. Acta Bot. Boreal. -Occident. Sin. 30(7):1388–1396.
- Pan SL, Shun QS, Bo QM, Bao XS. 2002. The coloured atlas of the medicinal plants from genus Bupleurum in China. Shanghai: Shanghai Science and Technology Literature Publishing House.
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15(1):8–15.
- She ML, Watson MF. 2005. Bupleurum Linnaeus. In Flora of China, Apiaceae through Ericaceae. Beijing, China: Science Press. p. 60–74.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang C, Ma X, He X. 2011a. Fruit features of some Bupleurum Species (Apiaceae) and their systematical implication. Plant Sci J. 29(4):399–408.
- Wang CB, Ma XG, He XJ. 2011b. A taxonomic re-assessment in the Chinese Bupleurum (Apiaceae): insights from morphology, nuclear ribosomal internal transcribed spacer, and chloroplast (trnH-psbA, matK) sequences. J Sys Evol. 49(6):558–589.
- Yang ZY, Chao Z, Huo KK, Xie H, Tian ZP, Pan SL. 2007. ITS sequence analysis used for molecular identification of the Bupleurum species from northwestern China. Phytomedicine. 14(6):416–423.
- Zhang GX. 2020. Survey and identification of cultivated Bupleurum spp. Germplasm resources in China. Ph.D. thesis. Peking Union Medical College, Beijing.