Abstract
In this study, the complete chloroplast genome of Michelia chartacea B. L. Chen & S. C. Yang was 160,138 bp in length. It includes a large single-copy (LSC) region of 88,164 bp, a small single-copy region (SSC) of 18,824 bp, and with a pair of inverted repeats (IRs) of 26,575 bp. The GC content in the chloroplast genome was 39.23%. In total, 130 genes in the chloroplast genome of Michelia chartacea were annotated, including 83 protein-coding genes, 38 tRNA genes, and eight rRNA genes. The phylogenetic analysis showed that M. chartacea was closely related with M. martini and M. maudiae, forming a clade included in Michelia.
The Magnoliaceae divided into several smaller genera is a family of lowering plant within the order Magnoliales (Law Citation1984; Liu Citation2004; Xia et al. Citation2009), and is considered as one of the most primitive groups of angiosperms (Li and Guo Citation2014). Michelia chartacea B. L. Chen & S. C. Yang (Michelia chapensis) belonging to the magnolia is a dominant tree species in the secondary evergreen broadleaf forests of China, it is even endemic to particular regions where background O3 concentrations exceed the thresholds (GB3095-2012) for forest protection in South China (Pan et al. Citation2020). Analyses of complete chloroplast genomes have the advantage to significantly improve the resolution of phylogenetic relationships in large, complex plant lineages (Doorduin et al. Citation2011). Due to the characteristics of uniparental inheritance, haploid nature, conserved structure and gene content, small genome size, chloroplast genomes have been widely applied in phylogenetic reconstructions (Dong et al. Citation2018) and molecular evolution (Walker et al. Citation2014). Here, the complete cp genome sequence of M. chartacea was assembled and analyzed using high-throughput sequencing technology, the annotated cpDNA has been deposited into GenBank with the accession number MT449723.
The fresh leaves were sampled in Longzhong Botanical Garden (32°10′N, 112°10′E), Hubei, China. The specimen of M. martini was stored in the herbarium from the Three Gorges University, the accession number was 15005121. The leaves from a single individual M. chartacea plant were rapidly frozen with liquid nitrogen and stored at −80 °C until used. Total genomic DNA was extracted using the modifed CTAB method to construct a library for sequencing with Illumina Hiseq 2500 platform (Illumina, San Diego, CA, USA). Additionally, MITObim v 1.8 (https://github.com/chrishah/MITObim) was used to assemble the complete circular cp genome sequence (Hahn et al. Citation2013). The cp genome was annotated and manually adjusted with CpGAVAS2 (http://www.herbalgenomics.org/cpgavas2) (Shi et al. Citation2019). The annotated sequence was submitted to NCBI. The genome sequence data that support the findings of this study are openly available in the GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. SUB8488805. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA675703, SRR13045767, and SAMN16711956, respectively.
The chloroplast genome of M. chartacea was a closed circular molecule of 160,138 bp presenting a typical quadripartite structure, of which the length of a large single-copy region (LSC) was 88,164 bp and the length of a small single-copy (SSC) region was 18,824 bp, which were separated by the IRA and IRB of 26,575 bp. The contents of CG in the chloroplast genome was 39.23%. A total of 130 chloroplast genes were annotated, containing 83 protein-coding genes (63.85%), 38 transporter RNA genes (29.23%), eight ribosomal RNA genes (6.15%), one pseudogene (0.77%) was inferred to be pseudogenes. Fifteen distinct genes (trnK-UUU, rps16, trnS-CGA, rpoC1, trnL-UAA, trnC-ACA, petB, rp12, ndhB, trnE-UUC, trnA-UGC, ycf1, ndhA, trnA-UGC and trnE-UUC) contained one intron, while ycf3 and clpP each contained two introns.
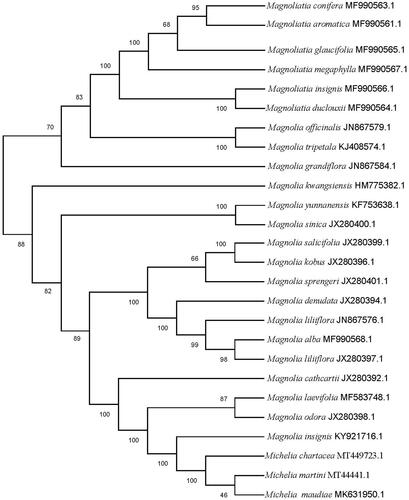
To estimate the putative performance of chloroplast genomes on their phylogenetic placements within the family Magnoliaceae, thus, the phylogenetic relationships was performed using the complete cp genomes of M. chartacea with those of obtained from 25 other species of Magnoliaceae reported in Genbank of NCBI database based on maximum likelihood (ML) analysis using MEGA 7.0 (Kumar et al. Citation2016) (https://www.megasoftware.net). The phylogenetic analysis showed that M. chartacea was closely related with M. martini and M. maudiae (Wang et al. Citation2019), forming a clade included in Michelia (). The cp genome of M. chartacea will provide important data for the further study of Magnoliaceae and systematics of the genus Michelia.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov/, reference number MT449723.
Additional information
Funding
References
- Dong W, Xu C, Wu P, Cheng T, Yu J, Zhou S, Hong D-Y. 2018. Resolving the systematic positions of enigmatic taxa: manipulating the chloroplast genome data of Saxifragales. Mol Phylogenet Evol. 126:321–330.
- Doorduin L, Gravendeel B, Lammers Y, Ariyurek Y, Chin-A-Woeng T, Vrieling K. 2011. The complete chloroplast genome of 17 individuals of pest species Jacobaea vulgaris: SNPs, microsatellites and barcoding markers for population and phylogenetic studies. DNA Res. 18(2):93–105.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-abaiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Law YW. 1984. A preliminary study on the taxonomy of the family Magnoliaceae. Acta Phytotaxon. Sin. 22(2):89–109.
- Li ZY, Guo R. 2014. Review on propagation biology and analysis on endangered factors of endangered species of Manglietia. Life Sci Res. 18:90–94.
- Liu YH. 2004. Woonyoungia law. In Liu Yuhu (Ed.), Magnolias of China. Beijing: Beijing Science and Technology Press; p. 381–383.
- Pan L, Zou XJ, Lie GW, Xue L, Chen HY. 2020. Ozone-induced changes in physiological and biochemical traits in Elaeocarpus sylvestris and Michelia chapensis in South China. Atmos Pollut Res. 11(5):973–980.
- Shi LC, Chen HC, Jiang M, Wang LQ, Wu X, Huang LF, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–73.
- Walker JF, Zanis MJ, Emery NC. 2014. Comparative analysis of complete chloroplast genome sequence and inversion variation in Lasthenia burkei (Madieae, Asteraceae)). Am J Bot. 101(4):722–729.
- Wang JQ, Li YY, Wang Q, Fan WW. 2019. Characterization of the complete chloroplast genome of Michelia maudiae (Magnoliaceae). Mitochondrial DNA Part B. 4(2):2146–2147.
- Xia NH, Liu YH, Nooteboom HP. 2009. Magnoliaceae. In Xia Nianhe, Liu Yuhu, Nooteboom HP. (Eds.), Flora of China. Vol. 7. Beijing: Missouri Botanical Garden Press and Science Press; p. 48.