Abstract
The complete chloroplast genomes of Trapa quadrispinosa and T. bicornis var. taiwanensis were reported in this study. The chloroplast genome of T. quadrispinosa was 155,554 bp in length, containing an LSC of 88,506 bp, an SSC of 18,274 bp, and a pair of IR regions of 24,387 bp each. The chloroplast genome of T. bicornis var. taiwanensis was 155,543 bp in length, including an LSC of 88,497 bp, an SSC of 18,274 bp, and a pair of IR regions of 24,386 bp each. Both genomes had 112 genes, consisting of 78 protein-coding genes, 30 tRNA genes, and four rRNA genes. The phylogenetic analysis revealed that the family Trapaceae was closely related to the family Sonneratiaceae.
Trapa L. is a genus with annual aquatic plants belonged to the family Trapaceae (Wu et al. Citation1999). The Amur River and Tumen River Basins and Yangtze River Basin are two biodiversity distribution centers of the genus Trapa around the world (Xue et al. Citation2016). Trapa has a high-level content of starch, with important medicinal and economic values. However, there still have many questions on the species delimitation and phylogenetic position of Trapa (Xue et al. Citation2017). In this study, the complete chloroplast genomes of Trapa quadrispinosa Roxb. and T. bicornis var. taiwanensis (Nakai) Z. T. Xiong were reported.
The materials of T. quadrispinosa and T. bicornis var. taiwanensis were collected from Liangzi Lake (30°26′N, 114°45′E, Hubei Province, China) and Hongze Lake (33°22′N, 118°42′E, Jiangsu Province, China), respectively. The specimens of them have been kept in Institute of Botany, Chinese Academy of Sciences (T20181006; T20181016). Total genomic DNA was extracted using the modified CTAB method (Doyle and Doyle Citation1987) and sequenced using the Illumina Hiseq Platform (Illumina Inc., San Diego, CA). Genomes were assembled by GetOrganelle v1.5 (Jin et al. Citation2018) and annotated using PGA (Qu et al. Citation2019).
The chloroplast genome of T. quadrispinosa (MT941481) was 155,554 bp in length with a GC content of 36.40%, containing a pair of IRs of 24,387 bp each which divide LSC of 88,506 bp and SSC of 18,274 bp. The chloroplast genome of T. bicornis var. taiwanensis (MT941480) was 155,543 bp in length with an overall GC content of 36.40%, including an LSC of 88,497 bp, an SSC of 18,274 bp, and a pair of IRs of 24,386 bp each. Both genomes had 112 genes, consisting of 78 protein-coding genes, 30 tRNA genes, and four rRNA genes.
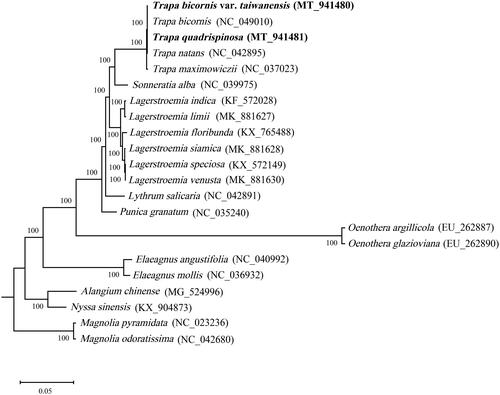
To confirm the phylogenetic position of T. quadrispinosa and T. bicornis var. taiwanensis among the Myrtales species, chloroplast genome sequences were aligned using MAFFT (Katoh and Standley Citation2013), and maximum-likelihood phylogenetic tree was constructed by MEGA 7.0 (Kumar et al. Citation2016) with 1000 bootstraps. The maximum-likelihood phylogenetic tree based on 22 complete chloroplast genomes, Magnolia pyramidata and M. odoratissima in the genus of Magnolia of Magnoliaceae family was used as outgroup. The results revealed that the family Trapaceae had a close relationship with the family Sonneratiaceae. T. quadrispinosa and T. bicornis var. taiwanensis were closely related to T. bicornis and T. natans with a low-level of divergence ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession nos. MT941481–MT941480. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA672284, SAMN16562397, and SAMN16562398, respectively.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. BioRxiv. 256479..
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Wu ZY, Raven PH, Hong DY. 1999. Flora of China. Vol. 53. Beijing, China: Beijing Science Press; p. 3–26.
- Xue ZQ, Xue JH, Kryukova MV, Ma KP. 2017. The complete chloroplast DNA sequence of Trapa maximowiczii Korsh. (Trapaceae), and comparative analysis with other Myrtales species. Aquatic Bot. 143:54–62.
- Xue JH, Xue ZQ, Wang RX, Rubtsova TA, Pshennikova LM, Guo YM. 2016. Distribution pattern and morphological diversity of Trapa L. in the Heilong and Tumen River Basin. Plant Sci J. 34(4):506–520.