Abstract
The complete chloroplast genome sequence of Artocarpus petelotii was determined, a narrowly distributed species at high altitudes in the family Moraceae. To better determine its phylogenetic location with respect to the other Moraceae species, the complete plastid genome of A. petelotii was sequenced. The whole chloroplast genome is 161,009 bp in length, consisting of a pair of inverted repeat (IR) regions of 25,682 bp, one large single-copy (LSC) region of 89,552 bp, and one small single-copy (SSC) region of 20,093 bp. The overall GC content of the whole chloroplast genome is 35.8%. Further, maximum likelihood phylogenetic analysis was conducted using 26 complete plastomes of the Moraceae, which support close relationships among A. petelotii, A. nanchuanensis and A. heterophyllus.
Artocarpus petelotii Gagnep is a member of the genus Artocarpus (Moraceae), which includes approximately 70 species mostly native to the hot regions of South and Southeast Asia and Oceania (William et al. Citation2017). Artocarpus petelotii is a narrowly distributed species at high altitudes in Southeast Yunnan of China and North Vietnam (Wu and Zhang Citation1989). The fruits of A. petelotii contains plenty of nutritional compositions with potential great medicinal value (Ren et al. Citation2014). For a better understanding of the relationships of A. petelotii and other Moraceae species, it is necessary to reconstruct a phylogenetic tree based on high-throughput sequencing approaches.
Fresh young leaves of A. petelotii in Xishuangbanna (Yunnan, China; Long. 101.2611 E, Lat. 21.9275 N, 555 m) were picked for DNA extraction (Doyle and Dickson Citation1987). The voucher was deposited at the Biodiversity Research Group of Xishuangbanna Tropical Botanical Garden (Accession Number: XTBG-BRG-10003). Long-range polymerase chain reaction was carried out following Zhang et al. (Citation2016), with their 15 pairs of universal primers. The 15 purified polymerase chain reaction products were mixed with 0.4 μg for each. A total of 6 μg product was sent to BGI, Shenzhen for library construction and genome sequencing on the Illumina Hiseq 2000 Platform (Illumina, San Diego, CA), more than 100 Mb of sequence data for the sample was obtained and subjected to chloroplast gnome assmembly. Paired-end reads were assembled using the trial version of CLC v.8 (http://www.qiagenbioinformatics.Com). The contigs were aligned using the publicly available plastid genome of Morus celtidifolia (Accession number MT154045). The complete chloroplast genome was annotated in Geneious 10.1.3 (Kearse et al., Citation2012).
The chloroplast genomes of A. petelotii (Accession number MW250918), with a length of 161,009 bp, was 257 bp, 622 bp, 1524 bp and 2550 bp larger than that of A. nanchuanensis (160,752 bp, LAU10104), A. heterophyllus (160,387 bp, MG434693), Morus celtidifolia (159,485 bp, MT154045) and M. mongolica (158,459 bp, KM491711). It was also 1161 bp smaller than that of Broussonetia kurzii (162,170 bp, MH118529). The length of the inverted repeats (IRs), large single-copy (LSC), and small single-copy (SSC) regions of A. petelotii was 25,682 bp, 89,552 bp, and 20,093 bp, respectively. The overall G + C content is 35.8% (LSC, 33.4%; SSC, 28.6%; IR, 42.8%).
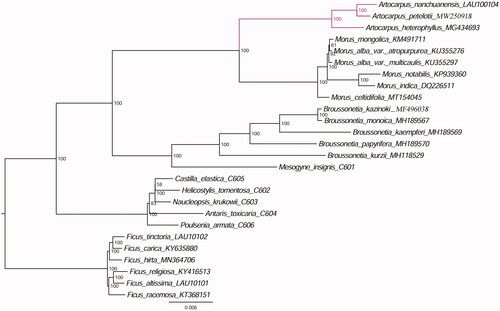
Furthermore, based on 16 previously reported plastomes from Genbank including A. heterophyllus, M. mongolica, M. alba var. Atropurpurea, M. alba var. multicaulis, M. notabilis, M. indica, M. celtidifolia, B. kazinoki, B. monoica, B. kaempferi, B. papyrifera, B. kurzii, Ficus carica, Ficus hirta, Ficus religiosa, Ficus racemosa, 3 plastome sequences reported in our previous study including A. nanchuanensis, Ficus tinctoria, Ficus altissima and 6 published plastome genome sequences (Bruun-Lund et al. Citation2017) including Mesogyne insignis, Castilla elastica, Helicostylis tomentosa, Naudeopsis krukowii, Antaris toxicaria, Poulsenia armata, we reconstructed a phylogenetic tree () with Ficus tinctoria, Ficus carica, Ficus hirta, Ficus religiosa, Ficus altissima, Ficus racemosa as outgroups. Maximum likelihood (ML) phylogenetic analysis were performed base on TVM + F+R2 model in the iqtree version 1.6.7 program with 1000 bootstrap replicates (Nguyen et al. Citation2015). The ML phylogenetic tree with 58–100% bootstrap values at each node supported that A. petelotii was closely related to A. nanchuanensis and A. heterophyllus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the finding of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW250918.
Additional information
Funding
References
- Bruun-Lund S, Clement WL, Kjellberg F, Rønsted N. 2017. First plastid phylogenomic study reveals potential cyto-nuclear discordance in the evolutionary history of Ficus L. (Moraceae). Mol Phylogenet Evol. 109:93–104.
- Doyle JJ, Dickson EE. 1987. Preservation of plant samples for DNA restriction endonuclease analysis. Taxon. 36(4):715–722.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Ren G, Yi WF, Zhong GY, Yuan WJ, Peng JB, Ma ZL, Zeng JX. 2014. Artostyracins A-C, three new isoprenylated 2-arylbenzofurans from Artocarpus styracifolius. Phytochem Lett. 10:235–239.
- William EW, Gardner EM, Harris R, Chaveerach A, Pereira JT, Zerega NJC. 2017. Out of Borneo: biogeography, phylogeny and divergence date estimates of Artocarpus (Moraceae). Ann Bot. 119:611–627.
- Wu ZY, Zhang XS. 1989. Taxa nova nonnulla Moracearum sinensium. Acta Bot Yunnanica. 11:24–34.
- Zhang T, Zeng CX, Yang JB, Li HT, Li DZ. 2016. Fifteen novel universal primer pairs for sequencing whole chloroplast genomes and a primer pair for nuclear ribosomal DNAs. J Sytemat Evol. 54(3):219–227.