Abstract
Plants in the genus Melaleuca have been widely used as traditional medicine mainly because of their broad spectrum antimicrobial activity. In this study, we reported the complete chloroplast genome of Melaleuca cajuputi subsp. cumingiana. The chloroplast genome of this species is 158,855 bp in length, including a pair of inverted repeat regions (IRs) (26,727 bp) that is divided by a large single-copy (LSC) area (87,338 bp) and a small single-copy (SSC) area (18,063 bp). The circular chloroplast genome of M. cajuputi subsp. cumingiana contains 135 unique genes, composing of 87 protein-coding genes, 40 tRNA genes, and eight rRNA genes. Phylogenetic analysis indicates that M. cajuputi subsp. cumingiana was clustered with species in the tribe Melaleuceae. This complete chloroplast genome of M. cajuputi subsp. cumingiana will provide a powerful tool to accelerate breeding, biotechnological and phylogenetic study.
Plants belonging to Melaleuca L. genus (Myrtaceae family) have been used as traditional medicine for many years (Sharifi-Rad et al. Citation2017), mainly because of their broad spectrum antimicrobial activity (Zhang et al. Citation2018). Melaleuca cajuputi Powell is a multi-purpose tree as its piles and frame poles provide construction materials, leaves produce essential oil, flowers attract honey bees (Doran and Turnbull Citation1997; Quat and Cuong Citation2005), and timbers can be used for pulp, fiber, and particle board (Trung Citation2008). In recent years, this plant was also developed for allelopathic herbicides instead of chemical herbicides (Kueh et al. Citation2019). There are three recognized subspecies: cajuputi, cumingiana, and platyphylla (Craven and Barlow Citation1997), which are native to Australia and adjacent areas such as Papua New Guinea, Indonesia, and Malaysia (Brophy et al. Citation2013). Breeding program of M. cajuputi subsp. cajuputi has mainly targeted essential oil yield (Kartikawati et al. Citation2015), while M. cajuputi subsp. cumingiana Barlow was widely planted for its wood production (Thiet et al. Citation2017; Nguyen et al. Citation2019). However, only a few genomic resources have been reported in this species (Beheregaray and Sunnucks Citation2000).
In higher plants, chloroplast genome is often used for phylogenetic analysis and domestication studies (Jansen et al. Citation2007). The whole chloroplast genome sequences also have demonstrated the potential to understand structure and functional evolution (Jansen et al. Citation2007; Moore et al. Citation2010). In genus Melaleuca, the chloroplast genome such as M. alternifolia and M. rigidus (which was initially described as a member of the genus Callistemon) has been reported (Liu et al. Citation2019, Citation2020), but the chloroplast genome of M. cajuputi has not been reported. Here, we sequenced and analyzed the complete chloroplast genome sequence of M. cajuputi subsp. cumingiana based on the Illumina sequencing data. The objective of this study was to characterize the complete chloroplast genome sequence of M. cajuputi as a resource for future genetic studies on this and other related species.
Voucher specimens of M. cajuputi subsp. cumingiana were collected from South China Botanical Garden, Chinese Academy of Sciences (Guangzhou, China; 113°21′7″E, 23°10′47″N), and deposited at the herbarium of South China Botanical Garden (accession number: SCBG-CF-2071). Total genomic DNA was extracted from fresh leaves using the CTAB-chloroform protocol (Doyle and Doyle Citation1987). The high-throughput sequencing (pair-end 150 bp) was performed on an Illumina XTen platform and it generated ∼3.07 Gb raw data. The cp genome was assembled by using the program NOVOPlasty (Dierckxsens et al. Citation2017). A ribulose-1, 5-bisphosphate carboxylase/oxygenase (rbcL) gene sequence from M. alternifolia (GenBank accession no. MN310606) was used as seed sequence, and the whole cp genome sequence of M. alternifolia and Eucalyptus grandis (GenBank accession no. NC_014570) was used as a reference to resolve the inverted repeat (IR) in the chloroplast genome of M. cajuputi subsp. cumingiana. The assembled chloroplast genome was annotated using PGA (Qu et al. Citation2019) and GeSeq (Tillich et al. Citation2017). For necessary genes, positions of start and stop codons and boundaries between exons and introns were manually corrected. The annotated chloroplast genomic sequence has been deposited in GenBank with an accession number: MT731621.
The complete chloroplast genome of M. cajuputi subsp. cumingiana is 158,855 bp in length, and has a typical quadripartite construction, which contains two inverted repeat regions (IRa and IRb) of 26,727 bp that is insulated by a large single-copy (LSC, 87,338 bp) and a small single-copy (SSC, 18,063 bp). The total GC content of complete chloroplast genome, LSC, SSC, IR regions is 36.8%, 55.0%, 11.4%, and 33.6%, respectively. The complete chloroplast genome of M. cajuputi subsp. cumingiana contains 135 unique genes, including 87 protein-coding genes, 40 tRNA genes, and eight rRNA genes. Introns are present in 19 of the annotated genes. Five of the intron containing genes contain three exons. Most of these genes are single-copy genes. However, 19 genes were duplicated in IR regions.
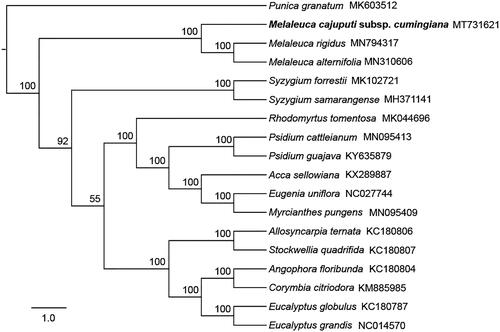
To confirm the phylogenetic position of M. cajuputi subsp. cumingiana, the complete chloroplast genomes of 16 published species within Myrtaceae and one outgroup (Punica granatum, Lythraceae, MK603512) were downloaded from the NCBI GenBank database. Ninety-four chloroplast genes shared by all species in this analysis were extracted, and were aligned by using MUSCLE (Edgar Citation2004). We concatenated these genes and then constructed a maximum-likelihood tree () using IQ-TREE (Nguyen et al. Citation2015). Phylogenetic analysis strongly supported that M. cajuputi subsp. cumingiana was closely related to species in tribe Melaleuceae (), which is consistent with the previous study in Myrtaceae (Thornhill et al. Citation2015). In addition, phylogenetic position of Melaleuca rigidus, initially described as Callistemon rigidus, also supported the sinking of Callistemon into Melaleuca (Craven Citation2006). In conclusion, we assembled the first chloroplast genome of M. cajuputi and it will provide a solid foundation for phylogenetic and evolutionary studies in Melaleuca and is expected to contribute to improving M. cajuputi breeding.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The contact person of specimen is Chen Feng ([email protected]). The raw sequencing data of M. cajuputi subsp. cumingiana have been deposited in the NCBI Sequence Read Archive under accession numbers PRJNA674705. The chloroplast genome of the M. cajuputi subsp. cumingiana was submitted to GenBank under accession number: MT731621. Treefile of 18 species and genes for phylogenetic analysis were deposited at Figshare: https://doi.org/10.6084/m9.figshare.13194128.v1.
Additional information
Funding
References
- Beheregaray LB, Sunnucks P. 2000. Microsatellite loci isolated from Odontesthes argentinensis and the O. perugiae species group and their use in other South American silverside fish. Mol Ecol. 9(5):629–644.
- Brophy JJ, Craven LA, Doran JC. 2013. Melaleucas: their botany, essential oils and uses. Australian Centre for International Agricultural Research (ACIAR) monograph no. 156. Canberra: CanPrint Communications.
- Craven LA. 2006. New combinations in Melaleuca for Australian species of Callistemon (Myrtaceae). Novon. 16(4):468–475.
- Craven LA, Barlow BA. 1997. New taxa and new combinations in Melaleuca (Myrtaceae). Novon. 7(2):113–119.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doran JC, Turnbull JW. 1997. Australian trees and shrubs: species for land rehabilitation and farm planting in the tropics. ACIAR Monograph No. 24; p. viii + 384.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 104(49):19369–19374.
- Kartikawati NK, Naiem M, Hardiyanto EB, Rimbawanto A. 2015. Improvement of seed orchard management based on mating system of cajuputi trees. Indonesian J Biotechnol. 18(1):26–35.
- Kueh BWB, Yusup S, Osman N, Ramli NH. 2019. Analysis of Melaleuca cajuputi extract as the potential herbicides for paddy weeds. Sustain Chem Pharm. 11:36–40.
- Liu H, Geng M, Qin Y, Xiao Y, Li M. 2019. Characterization of the complete chloroplast genome of medicinal tea tree (Melaleuca alternifolia). Mitochondrial DNA Part B. 4(2):3307–3308.
- Liu F, Movahedi A, Yang W, Xu L, Xie J, Zhang Y. 2020. The complete chloroplast genome and characteristics analysis of Callistemon rigidus R.Br. Mol Biol Rep. 47(7):5013–5024.
- Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci USA. 107(10):4623–4628.
- Nguyen THH, Rimbawanto A, Prastyono P, Kartikawati NK, Wu H. 2019. Genetic improvement for essential oil yield and quality in Melaleuca cajuputi. Ind Crop Prod. 137:681–686.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Quat NX, Cuong NV. 2005. Vietnamese Melaleuca-multi-ecology and multi-purpose tree species. Hanoi: Forest Science Institute of Vietnam.
- Sharifi-Rad J, Salehi B, Varoni EM, Sharopov F, Yousaf Z, Ayatollahi SA, Kobarfard F, Sharifi-Rad M, Afdjei MH, Sharifi-Rad M, et al. 2017. Plants of the Melaleuca genus as antimicrobial agents: from farm to pharmacy. Phytother Res. 31(10):1475–1494.
- Thiet NV, Dai DP, Lan PTM. 2017. Melaleuca seed orchards in long an: growth and potential for tree improvement. Vietnam Sci J. 2017:40–50.
- Thornhill AH, Ho SY, Kulheim C, Crisp MD. 2015. Interpreting the modern distribution of Myrtaceae using a dated molecular phylogeny. Mol Phylogenet Evol. 93:29–43.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Trung NQ. 2008. Melaleuca Timber- Resource Potential and Its Current Use in Kien Giang Province. Project “Conservation and Development of the Biosphere Reserve of Kien Giang Province. Technische Zusammenarbeit (GTZ) GmbH 27 pp.
- Zhang X, Guo Y, Guo L, Jiang H, Ji Q. 2018. In vitro evaluation of antioxidant and antimicrobial activities of Melaleuca alternifolia essential oil. Biomed Res Int. 2018:2396109.