Abstract
Holly (Ilex L.) is a woody dioecious genus cultivated as pharmaceutical, ornamentals, and industrial materials. Ilex suaveolens (H. Lév.) Loes is an endemic medicinal holly with a predominant distribution in Mount Huangshan, China. In the present work, the complete plastid genome of I. suaveolens was de novo sequenced by high-throughput sequencing technology. The newly-assembled plastid genome holds 37.6% of the overall GC content and a length of 157,857 bp, comprising a large single-copy (LSC, 87,255 bp), a small single-copy (SSC, 18,398 bp), and a pair of inverted repeat (IRs, 26,102 bp) regions. The plastid genome annotation suggested the presence of a total of 89 protein-encoding genes, 37 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. The plastome-mediated phylogenetic topology revealed that I. suaveolens clustered together with I. szechwanenesis and I. viridis in the same clade, and a strong relationship between clades and biogeography was found. These data contribute to the understanding of genetic diversity and conservation study of Ilex in Mount Huangshan.
Holly (Ilex L.), in the monogeneric family of Aquifoliaceae, is a living woody dioecious angiosperm genus, accounting for approximately 700 species (Yao et al. Citation2016). The majority of Ilex genus is used widely due to pharmaceutical, culinary, ornamental, and industrial materials. Approximately 204 Ilex species (149 endemic species) have been documented in the China Flora. Given that some systematic studies revealed a high incongruity of phylogenies with the traditional taxonomy, the evolutionary patterns remain to be explored further in Ilex (Manen et al. Citation2002; Yao et al. Citation2020).
In a transition zone of north-south flora of Eastern China, Mount Huangshan is regarded as a priority spot for biodiversity and conservation. Recent surveys prompted that more than 20 Ilex species displayed diversified medicinal properties and economic values (Hao et al. Citation2013; Qian and Tian Citation2016; Yi et al. Citation2016). Among them, I. suaveolens is the most proliferous local holly, exhibiting potential functions in scavenging heat, anti-inflammation, and detoxification (Ding et al. Citation2016). Here, the complete plastid genome of I. suaveolens was sequenced, providing essential data for taxonomy and conservation genetics and clues to explore new molecular markers among taxa in Aquifoliaceae (Nock et al. Citation2011; Zong et al. Citation2019).
The fresh leaves of I. suaveolens were harvested from a 10.24 ha (320 m × 320 m) forest plot (30°8′ 26″ N, 118°6′ 38″ E) in Mount Huangshan (Anhui, China). The dynamic plot ranges in altitude from 430 to 565 m with an annual average temperature of 7.8 °C and annual precipitation of 2394.5 mm. The voucher specimen (YL20190417016) was preserved in the herbarium of Nanjing Forestry University. DNA extraction was conducted according to a previous report (Su et al. Citation2019), and the next-generation sequencing of the whole-plastid genomes was served by Biodata Biotechnologies Inc. (Hefei, China) on the BGISEQ-500 platform (Shenzhen, China). Approximately 50 MB of high-quality clean paired-end reads was generated, followed by assembling the filtered sequences using SPAdes assembler 3.14.1 software (Bankevich et al. Citation2012). The plastid genome sequences were further annotated using the DOGMA (Wyman et al. Citation2004).
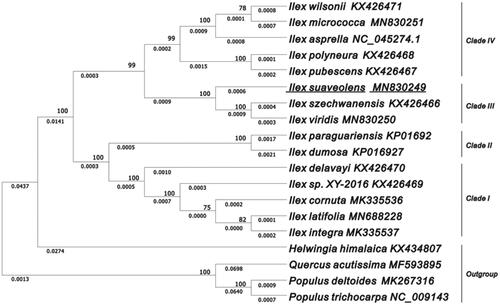
The plastid genome of I. suaveolens comprised a double-stranded and circular DNA (157,857 bp), containing two inverted repeat (IRs, 26,102 bp) regions separated by a large single-copy (LSC, 87,255 bp) and a small single-copy (SSC, 18,398 bp) sections. The overall GC content is 37.6%, and the corresponding values in LSC, SSC, and IRs regions are 35.7%, 31.9%, and 42.9%, respectively. The plastid genome was predicted to encode 134 genes, including 89 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Seven protein-coding genes, eight tRNA genes, and four rRNA genes show duplications in IR regions. Nineteen genes were identified to have two exons, and two genes (clpP and ycf3) contained three exons. Using MAFFT v7.471, the multiple sequences of 15 Ilex species were aligned (Katoh et al. Citation2019). The plastid topology of phylogenies was reconstructed using the software MEGA X, showing that I. suaveolens is mostly related to I. szechwanensis and I. viridis in the clade III (). In summary, the plastid phylogenetic tree displayed superior resolution for species discrimination and a better indication of the phylogeographic distribution in Ilex.
Figure 1. The evolutionary tree was constructed by MEGA X using the Maximum Likelihood method and the Tamura-Neighbour model. The percentage of phylogenetic trees associated with taxa is shown next to the branches. The analyses involved 15 plastomes of Ilex species with P. trichocarpa, P. deltoides, Q. acutissima, and H. himalaica rooted as the outgroup. The bootstrap values are shown on the branches of the phylogenetic tree based on 1000 replicates.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete plastid genome data that support the findings of this study are openly available in the GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number of MN830249. The raw sequence reads have been deposited in GSA database (https://bigd.big.ac.cn/gsa/) associated with the accession number of CRR147931.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Ding H, Fang Y, Yang X, Yuan F, He L, Yao J, Wu J, Chi B, Li Y, Chen S, et al. 2016. Community characteristics of a subtropical evergreen broad-leaved forest in Huangshan, Anhui Province, East China. Biodivers Sci. 24(8):875–887.
- Hao D, Gu X, Xiao P, Liang Z, Xu L, Peng Y. 2013. Research progress in the phytochemistry and biology of Ilex pharmaceutical resources. Acta Pharm Sin B. 3(1):8–19.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Manen JF, Boulter MC, Naciri-Graven Y. 2002. The complex history of the genus Ilex L. (Aquifoliaceae): evidence from the comparison of plastid and nuclear DNA sequences and from fossil data. Plant Syst Evol. 235(1):79–98.
- Nock CJ, Waters DLE, Edwards MA, Bowen SG, Rice N, Cordeiro GM, Henry RJ. 2011. Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol J. 9(3):328–333.
- Qian Y, Tian R. 2016. Research advance of Ilex germplasm resources and their application to landscape. World Forest Res. 29:40–45.
- Su T, Han M, Min J, Cao D, Pan H, Liu Y. 2019. The complete chloroplast genome sequence of Populus deltoides 'Siyang-2'. Mitochondrial DNA Part B. 5(1):283–285.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yao X, Song Y, Yang J, Tan Y, Corlett RT. 2020. Phylogeny and biogeography of the hollies (Ilex L., Aquifoliaceae). J Syst Evol. 00 (0): 1–10.
- Yao X, Tan Y-H, Liu Y-Y, Song Y, Yang J-B, Corlett RT. 2016. Chloroplast genome structure in Ilex (Aquifoliaceae). Sci Rep. 6:28559.
- Yi F, Zhao X, Peng Y, Xiao P. 2016. Genus Ilex L. phytochemistry, ethnopharmacology, and pharmacology. Chinese Herb Med. 8(3):209–230.
- Zong D, Gan P, Zhou A, Zhang Y, Zou X, Duan A, Song Y, He C. 2019. Plastome sequences help to resolve deep-level relationships of Populus in the family Salicaceae. Front Plant Sci. 10:5.
