Abstract
Six circular mitochondrial genomes of multi-, bi-, and uninucleate Rhizoctonia isolates were assembled and found that all the genomes contain 14 conserved protein-coding genes, one ribosomal protein (rps3), and 23 tRNA in the same order. The mitogenome sizes of uninucleate isolates were relatively smaller than binucleate and multinucleate stains. The size variations between uninucleate and multinucleate isolates were from both intergenic and intronic regions, whereas the differences between uninucleate and binucleate isolates were predominantly from intergenic regions. The phylogenetic analysis revealed that Rhizoctonia strains of the same nucleate types had a closer relationship.
Keywords:
The basidiomycetous fungal genus Rhizoctonia is composed of multinucleate, binucleate, and uninucleate isolates by the number of nuclei in the hyphal cells, in which the multinucleate R. solani is most popular and found to cause severe damage to more than 200 plant species (Sneh et al. Citation1991; Fang et al. Citation2013). With increasing numbers of binucleate and uninucleate isolates discovered, they are also reported to cause diseases on various plants (Lilja and Rikala Citation2000; Yang et al. Citation2014). Genomes of five multinucleate Rhizoctonia isolates have been analyzed with two mitogenomes presented (Wibberg et al. Citation2013; Losada et al. Citation2014). However, little is known about the mitogenomes of binucleate and uninucleate Rhizoctonia. In this study, we report six complete mitochondrial genomes of multi-, bi-, and uninucleate Rhizoctonia, isolated from maize and wheat, in which multinucleate Rhizoctonia isolate XN was more aggressive against maize than binucleate or uninucleate isolate, and provide a phylogenetic relationship of related taxa based on concatenated genes.
Six Rhizoctonia isolates with three uninucleate (JN, SM, and YR), two binucleate (LY and RW), and one multinucleate (XN) have been described (Li et al. Citation2011, Citation2014; Zhou et al. Citation2015, Citation2016) and collected in this laboratory at China Agricultural University (40.02 N, 116.28E). Whole-genome sequencing was performed using the Illumina HiSeq2000 system by BGI, Shenzhen, China. After filtering adapter and low-quality reads using Trimmomatic v0.3264 (Bolger et al. Citation2014), high quality reads matched to reference mitochondrial genomes of R. solani 7/3/14 (Wibberg et al. Citation2013) and Rhs1AP (Losada et al. Citation2014) were collected and further cleaned with Bowtie2 (Langmead and Salzberg Citation2012) to remove possible contaminant nuclear sequence. Mitochondrial genome was assembled using SPAdes v3.9.0 (Bankevich et al. Citation2012) and GapCloser_v1.12 (Luo et al. Citation2012) for gap filling and annotated using MFannot tool (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl).
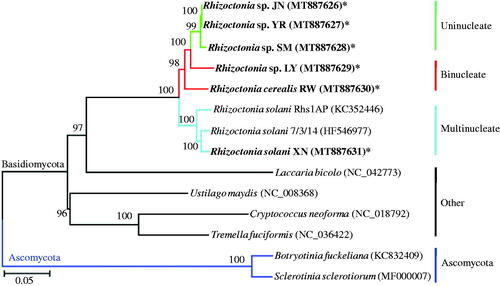
The sizes and GC contents of the six complete circular mitochondrial genomes were 126,087 bp and 39.66% for JN, 109,017 bp and 39.60% for SM, 120,576 and 39.23% for YR, 161,557 and 38.85% for LY, 153,061 and 39.64% for RW, and 151,045 bp and 33.84% for XN. The mitogenome sizes of uninucleate JN, SM, and YR were relatively smaller than binucleate LY and RW and multinucleate XN, whereas the GC content of XN was the lowest among the data set. The size differences between uninucleate and binucleate isolates were predominantly from intergenic regions, whereas the differences between uninucleate and multinucleate isolates were contributed from both intergenic and intronic regions. The gene composition and order were maintained across the six mitogenomes, including three subunits of ATP synthases (atp6, atp8, and atp9), apocytochrome b (cob), three subunits of cytochrome oxidases (cox1-3), seven subunits of NADH dehydrogenases (nad1-6 and nad4L), ribosomal protein (rps3), and 23 tRNAs, which were located on the same strand. A neighbor-joining phylogenetic relationship tree was constructed using MEGA 6.0 (Tamura et al. Citation2013) based on 15 mitochondrial proteins mentioned above, including R. solani Rhs1AP (Locus KC352446, Losada et al. Citation2014) and 7/3/14 (Locus HF546977, Wibberg et al. Citation2013), Ustilago maydis (Locus NC_008368), Cryptococcus neoforma (Locus NC_018792), Tremella fuciformis (Locus NC_036422), Laccaria bicolo (Locus NC_042773), and Ascomycetous Botryotinia fuckeliana (Locus KC832409) and Sclerotinia sclerotiorum (Locus MF000007) as outgroups. Rhizoctonia strains of the same nucleate types were grouped into the same subclade (), suggesting a closer relationship of the same nucleate isolates. The mitogenomes of multi-, bi-, and uninucleate Rhizoctonia provided an important information for understanding the evolution of Rhizoctonia.
Figure 1. Phylogenetic relationship between Rhizoctonia with other selected Basidiomycetous and Ascomycetous fungi. Fifteen mitochondrial proteins (atp6, atp8, atp9, cob, cox1-3, nad1-6, nad4L, and rps3) are used for Neighbour-Joining analysis using MEGA 6.0. The strains sequenced in this study were in bold and marked with “*”.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data of Rhizoctonia JN, YR, SM, LY, RW, and XN that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT887626-MT887631. The associated SRA numbers are SRR11566340, SRR13266617, SRR13296784, SRR11566450, SRR13258782, and SRR11560048, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kuliko AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Fang X, Finnegan PM, Barbetti MJ. 2013. Wide variation in virulence and genetic diversity of binucleate Rhizoctonia isolates associated with root rot of strawberry in Western Australia. PLoS One. 8(2):e55877.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359.
- Li W, Sun H, Deng Y, Zhang A, Chen H. 2014. The heterogeneity of the rDNA-ITS sequence and its phylogeny in Rhizoctonia cerealis, the cause of sharp eyespot in wheat. Curr Genet. 60(1):1–9.
- Li J, Xia HB, Yu F. 2011. The anastomosis groups of the corn sheath blight pathogen Rhizoctonia spp. in northeastern China. Mycosystema. 30:392–399.
- Lilja A, Rikala R. 2000. Effect of uninucleate Rhizoctonia on Scots pine and Norway spruce seedlings. Forest Pathol. 30(2):109–115.
- Losada L, Pakala SB, Fedorova ND, Joardar V, Shabalina SA, Hostetler J, Pakala SM, Zafar N, Thomas E, Rodriguez-Carres M, et al. 2014. Mobile elements and mitochondrial genome expansion in the soil fungus and potato pathogen Rhizoctonia solani AG-3. FEMS Microbiol Lett. 352(2):165–173.
- Luo RB, Liu BH, Xie YL, Li ZY, Huang WH, Yuan JY, He GZ, Chen YX, Pan Q, Liu YJ, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1(1):18.
- Sneh B, Burpee L, Ogoshi A. 1991. Identification of Rhizoctonia species. St. Paul, MN: APS Press; p. 519–520.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Wibberg D, Jelonek L, Rupp O, Hennig M, Eikmeyer FG, Goesmann A, Hartmann A, Borriss R, Grosch R, Pühler A, et al. 2013. Establishment and interpretation of the genome sequence of the phytopathogenic fungus Rhizoctonia solani AG1-IB isolate 7/3/14. J Biotechnol. 167(2):142–155.
- Yang Y, Zhao C, Guo Z, Wu X. 2014. Anastomosis groups and pathogenicity of binucleate Rhizoctonia isolates associated with stem canker of potato in China. Eur J Plant Pathol. 139(3):535–544.
- Zhou S, Liu Y, Zhang M, Li B, Chen X, Liang W. 2016. Comparison of the virulence and cognate virulence factors of multinucleate, binucleate and uninucleate Rhizoctonia isolates, causing sheath blight on maize plants. Eur J Plant Pathol. 145(2):501–506.
- Zhou S, Zhang M, Liu Y, Zhen J, Liang W, Chen X, Guo Z, Li B. 2015. A uninucleate Rhizoctonia sp. from maize plant with ITS heterogeneity and hypersensitive to abiotic stresses. Eur J Plant Pathol. 142(2):397–401.
