Abstract
The complete mitochondrial genome of an important medicinal plant Glycyrrhiza uralensis Fisch. is reported for the first time. The mitochondrial genome sequence of G. uralensis was 463,869 bp in length and had a GC content of 45.19%. The genome contained 40 protein-coding genes (PCGs), 30 transfer RNAs (tRNAs), and three ribosomal RNAs (rRNAs). The phylogenetic tree was built based on 25 plants, using the maximum-likelihood method. These data will provide certain help to determine the taxonomic status of G. uralensis.
Glycyrrhiza uralensis Fisch. is a perennial herb belonging to family Leguminosae, widely distributed from Central Asia to Northeast China. G. uralensis was first published in ‘Shen Nong's Materia Medica’, which has a history of 2000 years. It is known as the ‘King of Traditional Chinese Medicine’ in China. Studies on the effective biological activity of G. uralensis include antitumor (Aipire et al. Citation2017), antiviral (Wang et al. Citation2013), anti-inflammatory (Yang et al. Citation2017), and antibacterial (Chen et al. Citation2019), etc. Among the numerous researches on G. uralensis, most of them have focused on its pharmacological analysis and component comparative analysis. At present, whole-genome sequencing and discussion of some key genes have also been carried out (Liu et al. Citation2015; Mochida et al. Citation2017).
A sample was collected from fresh leaves of G. uralensis in Dalian, China (E 121°87′74.06″, N 39°06′18.21″). The G. uralensis plant samples and genomic DNA were stored in the herbarium of Liaoning University of Traditional Chinese Medicine (G. uralensis number: 10162200520025LY). G. uralensis genome was sequenced using a combination of the Nanopore platform (PromethION, Oxford Nanopore Technologies, Oxford, UK) and Illumina NovaSeq platform (Illumina, San Diego, CA). First, we used ABySS v2.0.2 (Simpson et al. Citation2009) to perform genome assembly with multiple-Kmer parameters and received the optimal results of the assembly. Second, BLASR (Chaisson and Tesler Citation2012) was used to map the preliminary assembly results to the Nanopore long reads. Then, SPAdes v3.10.1 (Bankevich et al. Citation2012) was used to assemble them together to construct contigs (Scaffolds), followed by error correction using Pilon v1.21 (Walker et al. Citation2014). The Nanopore assembled sequences were then checked if the sequence has overlapped and connected between them. The mitochondrial genome information of G. uralensis will provide data for further analysis of evolutionary history.
The mitochondrial genome of G. uralensis was sequenced by using the Illumina platform and Nanopore platform, which generated 26.5 million reads and 4.0 Gb raw data, respectively. Finally, the complete mitochondrial genome of G. uralensis was a circular form of 463,869 bp with an average read coverage of 3753× (coverage of Illumina reads was 3641.6×, coverage of Nanopore reads was 112.2×), and had a GC content of 45.19%. The mitochondrial genome encoded 73 unique genes, including 40 protein coding genes and 33 protein non-coding genes. The total length of the protein coding genes was 34,074 bp, accounting for 7.35% of the total length of the genome. The total length of non-coding proteins was 5129 bp (1.61%), including 30 transfer RNA (tRNA) genes and three ribosomal RNA (rRNA) genes (rrn18, rrn5, and rrn26). In addition, we found eight genes with introns (ccmFc, nad5, rps3, rps10, nad1, nad7, nad2, and nad4) containing 22 introns in total.
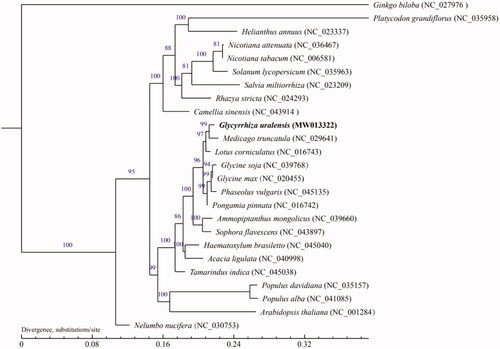
In order to understand the phylogenetic location of G. uralensis, we downloaded the mitochondrial genomes of 24 other plants and G. uralensis in NCBI to construct the phylogenetic tree with Ginkgo biloba as the outgroup. We use the MUMmer (Marcais et al. Citation2018) and BLAT software (Kent Citation2002) to do global alignment and local alignment between sample sequence and the reference genome under default parameters, and then manually optimized. The maximum-likelihood (ML) methods were performed for the genome-wide phylogenetic analyses using PhyML v3.0 (Guindon et al. Citation2010). Nucleotide substitution model selection was estimated with jModelTest v2.1.10 (Darriba et al. Citation2012) and Smart Model Selection in PhyML. The model GTR + I+G was selected for ML analyses with 1000 bootstrap (BS) replicates to calculate the BS values of the topology. The results tree was treated with iTOL v3.4.3 (Letunic and Bork Citation2016). It was found that G. uralensis and Medicago truncatula were closely clustered together (). At present, the available mitochondrial genome of angiosperm is still limited, which hinders our comprehensive understanding of the evolution of angiosperm mitochondrial genome. Besides the sequence level, plant mitochondrial genome can provide phylogenetic information at the structural level. These data will provide a basis for us to analyze the phylogenetic position of G. uralensis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW013322. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA679165, SRX9530379, and SRS7737028, respectively.
Additional information
Funding
References
- Aipire A, Li JY, Yuan PF, He J, Hu YL, Liu L, Feng XL, Li YJ, Zhang FC, Yang JH, et al. 2017. Glycyrrhiza uralensis water extract enhances dendritic cell maturation and antitumor efficacy of HPV dendritic cell-based vaccine. Sci Rep. 7:43796.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chaisson MJ, Tesler G. 2012. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics. 13:238.
- Chen YD, Agnello M, Dinis M, Chien KC, Wang J, Hu W, Shi WY, He XS, Zou J. 2019. Lollipop containing Glycyrrhiza uralensis extract reduces Streptococcus mutans colonization and maintains oral microbial diversity in Chinese preschool children. PLoS One. 14(8):e0221756.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12(4):656–664.
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245.
- Liu YL, Zhang PF, Song ML, Hou JL, Qing M, Wang WQ, Liu CS. 2015. Transcriptome analysis and development of SSR molecular markers in Glycyrrhiza uralensis Fisch. PLOS One. 10(11):e0143017.
- Marcais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. 2018. MUMmer4: a fast and versatile genome alignment system. PLOS Comput Biol. 14(1):e1005944.
- Mochida K, Sakurai T, Seki H, Yoshida T, Takahagi K, Sawai S, Uchiyama H, Muranaka T, Saito K. 2017. Draft genome assembly and annotation of Glycyrrhiza uralensis, a medicinal legume. Plant J. 89(2):181–194.
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19(6):1117–1123.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9(11):e112963.
- Wang JJ, Chen XQ, Wang W, Zhang YT, Yang ZY, Jin Y, Ge HM, Li EG, Yang G. 2013. Glycyrrhizic acid as the antiviral component of Glycyrrhiza uralensis Fisch. against coxsackievirus A16 and enterovirus 71 of hand foot and mouth disease. J Ethnopharmacol. 147(1):114–121.
- Yang R, Yuan BC, Ma YS, Zhou S, Liu Y. 2017. The anti-inflammatory activity of licorice, a widely used Chinese herb. Pharm Biol. 55(1):5–14.