Abstract
Two mitogenomes of long-tailed giant rat, Leopoldamys sabanus (Thomas, 1887), which belongs to the family Muridae were sequenced and assembled in this study. Both mitogenomes have a length of 15,973 bp and encode 13 protein-coding genes (PCGs), 22 transfer RNA genes, two ribosomal RNA genes and one control region. The circular molecule of L. sabanus has a typical vertebrate gene arrangement. Phylogenetic and BLASTn analysis using 10 Leopoldamys species mitogenomes revealed sequence variation occurred within species from different time zones. Along with the taxonomic issues, this suggests a landscape change might influence genetic connectivity.
The long-tailed giant rat, Leopoldamys sabanus is the common, generalist species in local assemblages of small mammals. This species is present throughout the Sunda region of Southeast Asia (Lim Citation1970) and has high mobility ranging between various forest matrices such as logged and unlogged forests (Wells et al. Citation2008). Recently, L. sabanus is reported to be widely distributed compared to the other non-volant mammals in northern forests of Peninsular Malaysia (Munian et al. Citation2020). In this study, we determined additional complete mitogenomes of L. sabanus from Malaysia that could be an important resource for addressing taxonomic issues and studying landscape genetics.
Leopoldamys sabanus sequenced in this study were collected from Bukit Tarek Forest Reserve, Selangor, Malaysia (3.48 N 101.47 E) (Faradiana et al. Citation2019) and Bukit Belate, Selangor, Malaysia (2.25 N 102.30 E). Total genomic DNA was extracted from the specimen tissues, which has been deposited at the Zoological Collection of Forest Research Institute Malaysia (FRIM) (Voucher No. MZF1958 and MZF731). The library was constructed using Blunt-End Single-Tube (BEST) protocol (Carøe et al. Citation2018). The mitogenome was assembled and annotated following Jahari, Abdul Malik, et al. (Citation2020) and Jahari, Mohd Azman, et al. (Citation2020). Both mitogenomes of L. sabanus (Genbank accession no. MT241668 and MT259591) have a length of 15,973 bp includes 13 protein-coding genes (PCGs), 22 transfer RNA genes, two ribosomal RNA genes and one control region.
These two L. sabanus mitogenomes display overall nucleotide composition which is 33.62% A, 28.68% T, 12.52% G and 25.17% C. The A + T content (62.30%) is higher than G + C content, which is similar to the other mitogenome of Leopoldamys species (Zhu et al. Citation2016; Camacho-Sanchez et al. Citation2017; Mohd Salleh et al. Citation2017). The total length of the protein-coding gene sequences (PCGs) is 11,405bp. The total length of the 22 tRNA genes is 1491bp, ranging from 57 bp (tRNASer) to 73 bp (tRNALeu). The 12S rRNA gene (955 bp) and 16S rRNA gene (1,573bp) are located between the tRNAPhe and tRNAVal, and between tRNAVal and tRNALeu, respectively. NAD6 and eight tRNAs genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu, tRNAPro) were encoded by the L-strand, other genes were encoded by the H-strand.
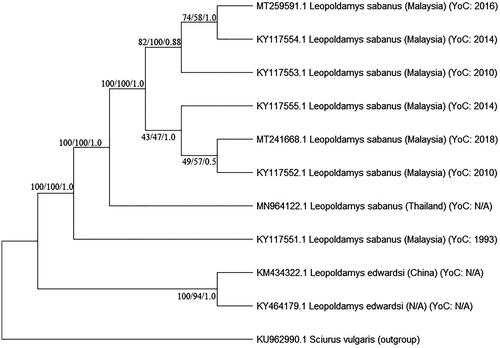
A phylogenetic tree of all available Leopoldamys mitogenomes was constructed using MEGA X software (Kumar et al. Citation2018). We confirmed that two L. sabanus in this study clustered with the other previously sequenced L. sabanus (Mohd Salleh et al. Citation2017; Nicolas et al. Citation2020) and rooted with the other Leopoldamys species (Zhu et al. Citation2016; Camacho-Sanchez et al. Citation2017) (). The comparison of these two newly sequenced mitogenomes to the Genbank using BLASTn found the closest match (more than 98% similarity) to the same species. However, it also showed sequence variation (92% similarity) when matched to the other sample of the same species (Genbank accession no. KY117551) (Mohd Salleh et al. Citation2017). Particularly, sample KY117551, which is a historical specimen collected from Sarawak instead of Peninsular Malaysia and has nearly a 30 years gap with the other L. sabanus in this study. The vicariance event which occurred in the Miocene and early Pliocene in Sunda Shelf, landscape variation and time lag factors could possibly alter the genetic connectivity between certain terrestrial species including Leopoldamys species (Gorog et al. Citation2004; Spear and Storfer Citation2008; Waits et al. Citation2016). In addition, it is also worth considering that L. sabanus may represent a complex of cryptic species due to the same morphology (Musser and Carleton Citation2005; Tamrin and Abdullah Citation2011). Thus, the mitogenomes generated and the analyses provided in this study address not only the taxonomic issues of the Leopoldamys species but also suggest further investigation on landscape genetics to examine how landscape change could influence genetic connectivity within Leopoldamys species.
Figure 1. The phylogenetic tree of L. sabanus mitogenomes (MT241668 and MT259591) and other Leopoldamys species available in Genbank. Bootstrap values were indicated in each branch of the tree representing the result of NJ/ML/Bayesian probability. Sciurus vulgaris was selected as outgroup (NA: not available; YoC: Year of Collection).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT241668 and MT259591. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA610427, SRR11241207 and SRR11241244, SAMN14297804 and SAMN14297815, respectively.
Additional information
Funding
References
- Camacho-Sanchez M, Leonard JA, Fitriana Y, Tilak M-K, Fabre P-H. 2017. The generic status of Rattus annandalei (Bonhote, 1903) (Rodentia, Murinae) and its evolutionary implications. J Mammal. 98(5):1340–1355.
- Carøe C, Gopalakrishnan S, Vinner L, Mak SST, Sinding MHS, Samaniego JA, Wales N, Sicheritz-Pontén T, Gilbert MTP. 2018. Single-tube library preparation for degraded DNA. Methods Ecol Evol. 9(2):410–419.
- Faradiana NMF, Shahfiz MA, Kaviarasu M, Nor Hazwani AR, Alwani NZ. 2019. Checklist on small vertebrates at Bukit Tarek Forest Reserve, Selangor. J Wildlife Parks. 34:119–128.
- Gorog AJ, Sinaga MH, Engstrom MD. 2004. Vicariance or dispersal? Historical biogeography of three Sunda shelf murine rodents (Maxomys surifer, Leopoldamys sabanus and Maxomys whiteheadi). Biol J Linnean Soc London. 81(1):91–109.
- Jahari PNS, Abdul Malik NF, Shamsir MS, Gilbert MTP, Mohd Salleh F. 2020. The first complete mitochondrial genome data of Hippocampus kuda originating from Malaysia. Data Brief. 31:105721.
- Jahari PNS, Mohd Azman S, Munian K, Ahmad Ruzman NH, Shamsir MS, Richter SR, Gilbert MTP, Mohd Salleh F. 2020. Molecular identification and phylogenetic analysis of a Callosciurus notatus complete mitogenome from Peninsular Malaysia. Mitochondrial DNA Part B. 5(3):3004–3024.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lim BL. 1970. Distribution, relative abundance, food habits, and parasite patterns of giant rats (Rattus) in West Malaysia. J Mammal. 51(4):730–740.
- Mohd Salleh F, Ramos-Madrigal J, Peñaloza F, Liu S, Mikkel-Holger SS, Riddhi PP, Martins R, Lenz D, Fickel J, Roos C, et al. 2017. An expanded mammal mitogenome dataset from Southeast Asia. GigaScience. 6(8):1–8.
- Munian K, Azman SM, Ruzman NA, Fauzi NFM, Zakaria AN. 2020. Diversity and composition of volant and non-volant small mammals in northern Selangor State Park and adjacent forest of Peninsular Malaysia. BDJ. 8:e50304.
- Musser G, Carleton M. 2005. Family Muridae. In: Wilson DE, Reeder DM, editors. Mammal species of the world: a taxonomic and geographic reference. 3rd ed. Baltimore (MD): Johns Hopkins University Press; p. 1189–1531.
- Nicolas V, Fabre P-H, Bryja J, Denys C, Verheyen E, Missoup AD, Olayemi A, Katuala P, Dudu A, Colyn M, et al. 2020. The phylogeny of the African wood mice (Muridae, Hylomyscus) based on complete mitochondrial genomes and five nuclear genes reveals their evolutionary history and undescribed diversity. Mol Phylogenet Evol. 144:106703.
- Spear SF, Storfer A. 2008. Landscape genetic structure of coastal tailed frogs (Ascaphus truei) in protected vs. managed forests. Mol Ecol. 17(21):4642–4656.
- Tamrin NAM, Abdullah MT. 2011. Molecular phylogenetics and systematics of five genera of Malaysian murine rodents (Maxomys, Sundamys, Leopoldamys, Niviventer and Rattus) inferred from partial mitochondrial cytochrome c oxidse subunit I (COI) gene. J Sci Technol Tropics. 7:75–86.
- Waits, L. P., Cushman, S. A., & Spear, S. F. (2016). Applications of landscape genetics to connectivity research in terrestrial animals. In N. Balkenhol, S. A. Cushman, A. T. Storfer, & L. P. Waits (Eds.), Landscape genetics: Concepts, methods, applications (pp. 199–219). Chichester, West Sussex, UK: Wiley-Blackwell.
- Wells K, Kalko EKV, Lakim MB, Pfeiffer M. 2008. Movement and ranging patterns of a tropical rat (Leopoldamys sabanus) in logged and unlogged rain forests. J Mammal. 89(3):712–720.
- Zhu D, Huang J, Kang C, Song X, Yue B, Zhang X. 2016. The complete mitochondrial genome of the Leopoldamys edwardsi (Rodentia: Muridae). Mitochondrial DNA Part A. 27(3):1882–1884.
