Abstract
This study was based on the collection of the complete genome of Lepidium perfoliatum chloroplast (cp). The full cp genome is 154,264 bp long, containing 130 genes, in which 8 genes are specified for ribosomal RNA (rRNA), while 85 and 37 genes for protein-coding and transfer RNA (tRNA) respectively. Phylogenetic analyss revealed the closed cluster of Lepidium perfoliatum with other Lepidium species such as Lepidium apetalum, Lepidium sativum, Lepidium meyenii and Lepidium virginicum, which helps for the evaluation of how Lepidium perfoliatum is phylogenetically related to other species.
Lepidium perfoliatum L. is a short-lived annual desert plant in the cruciferous family, naturally distributed in Xinjiang, Liaoning, Jiangsu, Gansu, and other places in China (Meng et al. Citation2008). Whole grass can be used as a medicine, with diuresis and anti-scurvy effect, etc. In addition, L. perfoliatum is also a kind of high-quality forage, with a unique physiological mechanism to adapt to the desert and early spring environment (Yang et al. Citation2015). In recent years, studies on the genus L. perfoliatum have focused on the characteristics of seed slime (Huang et al. Citation2015), total protein extraction (Ding et al. Citation2017), endophytic bacteria (Li et al. Citation2017), and salt tolerance (Roghieh et al. Citation2018). The genus Lepidium has been a relatively specialized group. Due to the lack of sufficient identification characteristics, the classification of genus Lepidium has always been difficult (Sun and Li Citation2007). However, no prior reports of the L. perfoliatum chloroplast genome are available, it may be important to elucidate the evolution of cruciferae species.
Fresh L. perfoliatum leaves were obtained from Carp mountain (Xinjiang, China, 87°34′716″E, 43°50′577″N) in Urumqi, Xinjiang, China and deposited the Voucher specimens in the Xinjiang Normal University herbarium (No. SHFZDXC02). Total extraction of gDNA was conducted via modified CTAB approach (Li et al. Citation2013) which was then utilized for building the Illumina pair-end library and its sequencing was carried out with an Illumina HiSeq platform (Illumina, CA, USA) at Tianjin Genomics Institute (TGS-Shenzhen, China) and yielded the raw data of approximately 4.4 GB. High-quality data contig assembly was then conducted with the SPAdes v3.9.0 de novo assembler (Bankevich et al. Citation2012), followed by futher assembly into an overall cp genome with NOVOPlast2.7.1 (Dierckxsens et al. Citation2017). Following assembly, these data were compared to the published L. sativum (MN176145) complete cp genome of. All cp gene annotation was conducted with the DOGMA tool using default parameters (Wyman et al. Citation2004).We have deposited the annotated cp genome of L. perfoliatum in Genbank under accession number MT880913.1.
The overall size of the L. perfoliatum cp genome is 154,264 bp with 36.46% GC content, which consists of 83,391 bp large single-copy region (LSC), 17,973 bp small single-copy region (SSC), and 26,450 bp inverted repeat regions pairs (IRA and IRB). It was predicted that the chloroplast genome consists of 130 unique genes, which includes 85 protein-coding genes, 8 rRNA, and 37 tRNA genes.
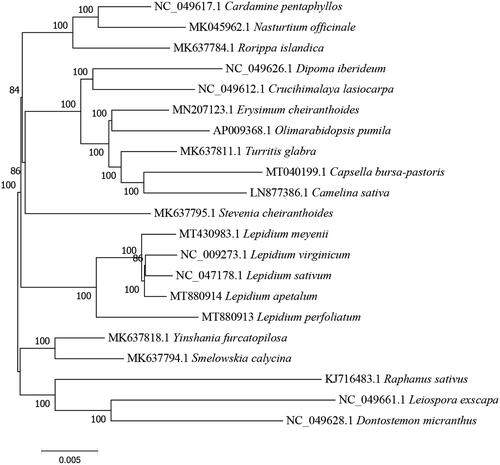
Of all the unique genes, 15 genes (trnK-UUU, trnL-UAA, rps16, trnG-UCC, atpF, trnV-UAC, rpoC1, ndhB, petB, rpl16, petD, rpl2, trnA-UGC, trnI-GAU, ndhA) contained a single intron, while 2 genes (clpP, ycf3) contained two introns. Similarly, one trans-splicing gene (rps12 gene) was also found. Phylogenic maximum-likelihood (ML) trees, based on a complete cp genome of 21 species which were obtained from the GenBank database, were constructed by using MAGE 7.0 (Kumar et al. Citation2016). Phylogenetic analysis indicated L. perfoliatum was sister to L. apetalum, L. sativum, L. meyenii, and L. virginicum, within Lepidium (), which is analogous to previously reported data (Zhu et al. Citation2019). Our study here could be further applied for the evolutionary and phylogenetic studies of this Brassicaceae Burnett plant.
Disclosure statement
The authors declare no conflicts of interest.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT880913.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA682689, SRR13213156, and SAMN16993569 respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Ding JP, Zhang N, Li Q. 2017. Comparative study of four methods of total protein extraction from seeds of two species of Lepidium Plants. Xinjiang Agric Sci. 54(07):1305–1312.
- Huang DH, Wang C, Yuan JW, Cao J, Lan HY. 2015. Differentiation of the seed coat and composition of the mucilage of Lepidium perfoliatum L.: a desert annual with typical myxospermy. Acta Biochim Biophys Sin (Shanghai). 47(10):775–787.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li YT, Cheng C, An DD. 2017. Characterisation of endophytic bacteria from a desert plant Lepidium perfoliatum L. Plant Prot Sci. 53(1):32–43.
- Li JL, Wang S, Yu J, Wang L, Zhou SL. 2013. A modified CTAB protocol for plant DNA extraction. Chin Bull Bot. 48(1):72–78.
- Meng J, Li Q, Li G. 2008. Physiological characteristic of seed germination of two species of Lepidium L. Biotechnology. 18(02):32–35.
- Roghieh H, Sara BR, Hossein A, Charlotte P. 2018. Salt tolerance mechanisms in three Irano-Turanian Brassicaceae halophytes relatives of Arabidopsis thaliana. J Plant Res. 131(6):1029–1046.
- Sun ZY, Li FZ. 2007. Studies on the leaf epidermal features of Lepidium (Brassicaceae) from China. Zhong Yao Cai. 30(7):780–785.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yang N, Zhao HP, Ge FW, Li YH, Zeng WJ, Zhao HX. 2015. Physiological response of two Lepidium species to low temperature stress during seed germination. Arid Zone Res. 32(4):760–765.
- Zhu B, Gao ZM, Luo X, Feng Q, Du XY, Weng QB, Cai MX. 2019. The complete chloroplast genome sequence of garden cress (Lepidium sativum L.) and its phylogenetic analysis in Brassicaceae family. Mitochondrial DNA B Resour. 4(2):3601–3602.