Abstract
Prunus fasciculata is a wild species of Prunus native to western North America. Here, we reported the complete chloroplast (cp) genome of P. fasciculata (GenBank accession number: MW160273). The cp genome was 157,986 bp long, with a large single-copy (LSC) region of 86,068 bp and a small single-copy (SSC) region of 19,166 bp separated by a pair of inverted repeats (IRs) of 26,376 bp. It encodes 129 genes, including 84 protein-coding genes, 37 tRNA genes, and eight ribosomal RNA genes. We also reconstructed the phylogeny of Prunus sensu lato using maximum-likelihood (ML) method, including our data and previously reported cp genomes of related taxa. The phylogenetic analysis confirmed the sister group relationship between P. fasciculata and the remaining subg. Prunus.
Desert almond, Prunus fasciculata (Torrey) A. Gray, is an intricately branched, deciduous, stiff stemmed shrub in Rosaceae that is widely scattered in and around the Mojave Desert of western North America (Mason Citation1913; Rohrer Citation2014). P. fasciculata is the representative species of Prunus subg. Prunus sect. Emplectocladus (Mason Citation1913) which had been described as a distinct genus (Torrey Citation1851) and then treated as a section within Prunus sensu lato (Gray Citation1874). A number of recent phylogenetic studies of Prunus sensu lato based on molecular data (Bortiri et al. Citation2001; Lee and Wen Citation2001; Wen et al. Citation2008; Shi et al. Citation2013; Chin et al. Citation2014) supported that the American section Emplectocladus diverged the earliest within subg. Prunus. The subg. Prunus is economically very important, but circumscription of subg. Prunus and its taxonomic status in Prunus sensu lato have long been controversial due to the still unsolved phylogenetic system of Prunus sensu lato (Shi et al. Citation2013; Chin et al. Citation2014). By taking advantages of next-generation sequencing technologies, we can rapidly access the abundant chloroplast (cp) genomic data for phylogenetic research (Li et al. Citation2017; Liu et al. Citation2017). Therefore, we sequenced the whole cp genome of P. fasciculata to elucidate its phylogenetic relationship with other species in Prunus sensu lato.
Total genomic DNA was extracted from silica-dried leaves collected from California Botanic Garden (Claremont, CA) using a modified CTAB method (Doyle and Doyle Citation1987). A voucher specimen (CALBG_6525) was collected and deposited in the Herbarium of Taizhou University. DNA libraries preparation and pair-end reads sequencing were performed on the Illumina NovaSeq 6000 platform (San Diego, CA). The cp genome was assembled via NOVOPlasty (Dierckxsens et al. Citation2017), using the Prunus rufa cp genome (MN648456; Li et al. Citation2020) as a reference. Gene annotation was performed via the online program Dual Organellar Genome Annotator (DOGMA; Wyman et al. Citation2004). Geneious R11 (Biomatters Ltd., Auckland, New Zealand) was used for inspecting the cp genome structure.
The complete cp genome of P. fasciculata (GenBank accession number: MW160273) was 157,986 bp long consisting of a pair of inverted repeat regions (IRs with 26,376 bp) divided by two single-copy regions (large single-copy (LSC) with 86,068 bp; small single-copy (SSC) with 19,166 bp). The overall GC contents of the total length, LSC, SSC, and IR regions were 36.7%, 34.6%, 30.1%, and 42.6%, respectively. The genome contained a total of 129 genes, including 84 protein-coding genes, 37 tRNA genes, and eight rRNA genes.
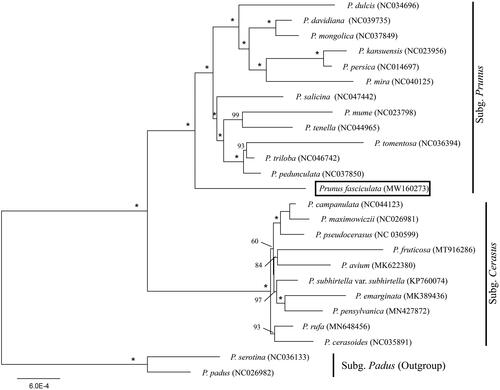
We used a total of 24 additional complete cp genomes of the Prunus sensu lato species to clarify the phylogenetic position of P. fasciculata. Prunus serotina Ehrh. (NC036133) and P. padus L. (NC026982) in Subg. Padus were used as the outgroup. We reconstructed a phylogeny employing the GTR + G model and 1000 bootstrap replicates under the maximum-likelihood (ML) inference in RAxML-HPC v.8.2.10 on the CIPRES cluster (Miller et al. Citation2010). The ML tree () was consistent with the most recent phylogenetic study on Prunus sensu lato (Shi et al. Citation2013; Chin et al. Citation2014). The phylogenetic analysis confirmed the sister group relationship between P. fasciculata and the remaining subg. Prunus.
Disclosure statement
The authors would like to thank California Botanic Garden for providing silica-dried leaves and voucher specimen of P. fasciculata. No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW160273. The associated BioProject, SRA, and BioSample numbers are PRJNA677866, SRR13039950, and SAMN16774453, respectively.
Additional information
Funding
References
- Bortiri E, Oh SH, Jiang JG, Baggett S, Granger A, Weeks C, Buckingham M, Potter D, Parfitt DE. 2001. Phylogeny and systematics of Prunus (Rosaceae) as determined by sequence analysis of ITS and the chloroplast trnL‐trnF spacer DNA. Syst Bot. 26:797–807.
- Chin SW, Shaw J, Haberle R, Wen J, Potter D. 2014. Diversification of almonds, peaches, plums and cherries – molecular systematics and biogeographic history of Prunus (Rosaceae). Mol Phylogenet Evol. 76:34–48.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Gray A. 1874. Contributions to the Botany of North America. In: American Academy of Arts and Sciences, editor. Proceedings of the American Academy of Arts and Sciences. Vol. 10. Boston: Press of John Wilson and Son. p. 39–78.
- Lee S, Wen J. 2001. A phylogenetic analysis of Prunus and the Amygdaloideae (Rosaceae) using ITS sequences of nuclear ribosomal DNA. Am J Bot. 88(1):150–160.
- Li YL, Clarke B, Li JH, Sun ZS. 2020. Complete chloroplast genome of Prunus rufa (Rosaceae), a wild flowering cherry endemic to the Himalayan region. Mitochondrial DNA Part B. 5:160–161.
- Li P, Lu RS, Xu WQ, Ohi-Toma T, Cai MQ, Qiu YX, Cameron KM, Fu CX. 2017. Comparative genomics and phylogenomics of East Asian Tulips (Amana, Liliaceae). Front Plant Sci. 8:451.
- Liu LX, Li R, Worth JRP, Li X, Li P, Cameron KM, Fu CX. 2017. The complete chloroplast genome of Chinese bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of Fagales. Front Plant Sci. 8:968.
- Mason SC. 1913. The pubescent-fruited species of Prunus of the southwestern states. J Agric Res. 1:147–177.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Comput Environ Workshop. 14:1–8.
- Rohrer JR. 2014. Prunus (Rosaceae). In: Flora of North America Editorial Committee, editors. Flora of North America North of Mexico. Vol. 9. New York: Oxford University Press. p. 352–383.
- Shi S, Li J, Sun J, Yu J, Zhou S. 2013. Phylogeny and classification of Prunus sensu lato (Rosaceae). J Integr Plant Biol. 55(11):1069–1079.
- Torrey J. 1851. On some new plants discovered by Col. Fremont, in California. Proc Am Assoc Adv Sci. 4:190–193.
- Wen J, Berggren ST, Lee CH, Ickert-Bond S, Yi TS, Yoo KO, Xie L, Shaw J, Potter D. 2008. Phylogenetic inferences in Prunus (Rosaceae) using chloroplast ndhF and nuclear ribosomal ITS sequences. J Syst Evol. 46:322–332.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.