Abstract
The complete mitochondrial genome of Syrphus ribesii was determined in this study. The double-stranded circular DNA molecule was 16,530 bp in length, containing 37 typical genes: 13 protein-coding genes (PCGs), 2 rRNA genes, 22 tRNA genes, and an A + T-rich region. Thirteen PCGs were 11,196 bp in size, encoding 3720 amino acids in total. All the PCGs started with ATN, except the COI used TTG as its initiation codon. Most PCGs terminated with standard codon TAA, while the COI ended with T and the ND5 ended with TA. The lrRNA and srRNA genes were 1341 bp and 793 bp in length, respectively. The A + T-rich region harbored some typical structures characteristic of the dipterans. The phylogenetic tree showed that Syrphus ribesii was closely related to Eupeodes corollae, and the Syrphidae and Pipunculidae constituted a monophyletic group within the Syrphoidea.
As one of the most abundant groups in Diptera, Syrphidae (Cyclorrhapha) is traditionally divided into three subfamilies (Eristalinae, Microdontinae, Syrphinae) covering about 230 genera, 6000 living species distributed nearly all around the world (Sommaggio Citation1999; Nedeljković et al. Citation2013). Most adults of Syrphidae have the habit of visiting flowers, making them one of the most important pollinators (Reemer Citation2013; Li Citation2019). However, to date, only about 20 complete mitochondrial genomes of Syrphidae have been determined (Pu et al. Citation2017; Li and Li Citation2020).
In this study, we determined the complete mitochondrial genome of Syrphus ribesii (Linnaeus, 1758), and conducted the phylogenetic analysis of the Syrphidae with other related groups. Adult individuals of Syrphus ribesii were collected from Shannan, Tibet Autonomous Region, China (91°76′E, 29°23'N) in July 2017. After sample collection, the fresh tissues were preserved in absolute ethanol immediately for fixation and stored at −20 °C in the Entomological Specimen Room of Anhui Normal University (Jiasheng Hao, [email protected]) under the voucher number ANUN-201707-1. The whole genomic DNA was extracted from thoracic muscle using the Animal Genome DNA Extraction Kit (Shanghai Sangon Biotech Co., Ltd., China), following the manufacturer’s protocols. The mitogenome of Syrphus ribesii was generated by amplified overlapping fragments using 3 pairs of universal PCR primers (Park et al. Citation2012) and a set of newly designed primers. PCR primers are available upon request. All fragments were sequenced on an ABI 3730XL DNA sequencer by Sangon Biotechnology Company (Shanghai, China) with primers walking on both strands. Sequences obtained were proofread and assembled using BioEdit v7.0.5 (Hall et al. Citation2011) and SeqMan program was included in the Lasergene software package (DNAStar, Inc., USA, NewYork). PCGs, rRNAs and the A + T-rich region were confirmed by the boundaries of tRNAs, and by alignment with other Syrphidae gene sequences using MEGA 7.0 (Kumar et al. Citation2016). The tRNA genes were identified by the MITOS2 WebServer (Bernt et al. Citation2013).
The complete mitogenome sequence of Syrphus ribesii was 16,530 bp in length (GenBank accession No. MW091497), including 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and an A + T-rich region. The gene arrangement and orientation were identical to other congeneric insects (Li et al. Citation2017; Chen et al. Citation2020). The base composition of the whole genome was detected to be 40.7% A, 40.4% T, 8.3% G and 10.7% C, exhibiting a relatively strong AT bias. Thirteen PCGs were 11,196 bp in size, encoding 3720 amino acids in total. All 13 PCGs started with the typical ATN codons, except for COI which was initiated by TTG. Eleven PCGs used universal TAA as their termination codons, whereas COI and ND5 used single T and TA as their stop codons, respectively. All tRNAs were well folded into a clover-leaf secondary structure except for tRNASer (AGN) losing the dihydrouridine (DHU) arm. The lrRNA and srRNA genes were 1341 bp and 793 bp in length, respectively. The A + T-rich region was 1491 bp in size, harboring some typical structures characteristic of dipterans: a 20 bp of poly-T stretch approximately in the center of the A + T-rich region and a microsatellite-like repeat (TA)9 located 127 bp upstream of the srRNA (Li et al. Citation2015; Zhang et al. Citation2016).
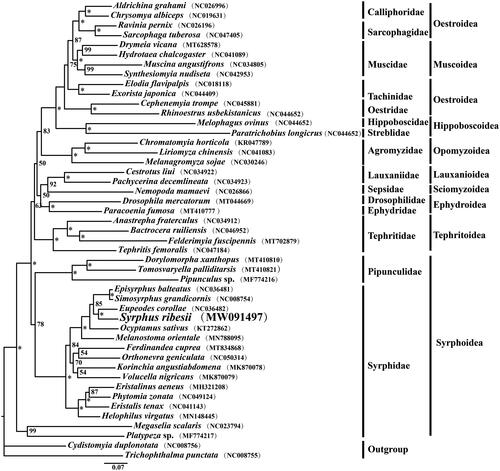
The phylogenetic analysis of the 45 Cyclorrhapha species was conducted based on the concatenated 13 PCG nucleotide sequence data with the maximum likelihood (ML) method, using two species from Tabanidae and Nemestrinidae as the outgroups (). The ML tree was inferred by IQ-TREE v1.6.8 (Nguyen et al. Citation2015) using the best-fitting GTR + I + G model which was estimated with ModelFinder (Kalyaanamoorthy et al. Citation2017) following the BIC criterion. The bootstrap support (BS) values of the tree nodes were calculated using 1000 replicates. The result indicated that Syrphus ribesii was closely related to Eupeodes corollae within the family Syrphidae. The Syrphidae and Pipunculidae formed a monophyletic group within the Syrphoidea, whereas the two families were shown to be paraphyletic in the previous study based on larger-scale transcriptomic data (Pauli et al. Citation2018).
Figure 1. The maximum-likelihood (ML) phylogenetic tree of 45 Cyclorrhapha species inferred from the concatenated 13 PCG nucleotide sequence data. The numbers on each tree node represents the bootstrap values (*BS = 100%). The alphanumeric characters in parentheses indicate the GenBank accession numbers.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI (National Center for Biotechnology Information) at https://www.ncbi.nlm.nih.gov/, reference number MW091497.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chen Q, Niu X, Fang Z, Weng Q. 2020. The complete mitochondrial genome of Melanostoma orientale (Diptera: Syrphidae). Mitochondrial DNA Part B. 5(1):554–555.
- Hall T, Biosciences I, Carlsbad C. 2011. BioEdit: an important software for molecular biology. GERF Bull Biosci. 2(1):60–61.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–591.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li XK, Ding SM, Cameron SL, Kang ZH, Wang YY, Yang D. 2015. The first mitochondrial genome of the sepsid fly Nemopoda mamaevi Ozerov, 1997 (Diptera: Sciomyzoidea: Sepsidae), with mitochondrial genome phylogeny of Cyclorrhapha. PLOS One. 10(3):e0123594.
- Li X, Ding S, Li X, Hou P, Tang C, Yang D. 2017. The complete mitochondrial genome analysis of Eristalis tenax (Diptera, Syrphidae). Mitochondrial DNA Part B. 2(2):654–655.
- Li H. 2019. Characterization and phylogenetic implications of the complete mitochondrial genome of Syrphidae. Genes. 10(8):563.
- Li J, Li H. 2020. The first complete mitochondrial genome of genus Phytomia (Diptera: Syrphidae). Mitochondrial DNA Part B. 5(3):2512–2513.
- Nedeljković Z, Ačanski J, Vujić A, Obreht D, Ðan M, Ståhls G, Radenković S. 2013. Taxonomy of Chrysotoxum festivum Linnaeus, 1758 (Diptera: Syrphidae) – an integrative approach. Zool J Linn Soc. 169(1):84–102.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Park JS, Cho Y, Kim MJ, Nam SH, Kim I. 2012. Description of complete mitochondrial genome of the black-veined white, Aporia crataegi (Lepidoptera: Papilionoidea), and comparison to papilionoid species. J Asia Pac Entomol. 15(3):331–341.
- Pauli T, Burt TO, Meusemann K, Bayless K, Donath A, Podsiadlowski L, Mayer C, Kozlov A, Vasilikopoulos A, Liu S, et al. 2018. New data, same story: phylogenomics does not support Syrphoidea (Diptera: Syrphidae, Pipunculidae). Syst Entomol. 43(3):447–459.
- Pu DQ, Liu HL, Gong YY, Ji PC, Li YJ, Mou FS, Wei SJ. 2017. Mitochondrial genomes of the hoverflies Episyrphus balteatus and Eupeodes corollae (Diptera: Syrphidae), with a phylogenetic analysis of Muscomorpha. Sci Rep. 7:44300
- Reemer M. 2013. Review and Phylogenetic evaluation of associations between Microdontinae (Diptera: Syrphidae) and ants (Hymenoptera: Formicidae). Psyche J Entom. 2013:1–9.
- Sommaggio D. 1999. Syrphidae: can they be used as environmental bioindicators? Agric Ecosyst Environ. 74(1–3):343–356.
- Zhang X, Kang ZH, Mao M, Li XK, Cameron SL, Jong H, Wang MQ, Yang D. 2016. Comparative mt genomics of the Tipuloidea (Diptera: Nematocera: Tipulomorpha) and its implications for the phylogeny of the Tipulomorpha. PLOS One. 11(6):e0158167.
