Abstract
The Bingzhi's stout newt (Pachytriton granulosus Chang, 1933) is distributed in mountainous areas of Zhejiang, China. The first complete mitochondrial genome (mitogenome) of P. granulosus was determined by next-generation sequencing. The size of the assembled mitogenome for P. granulosus was 16,293 bp, which included 13 protein coding genes, 22 tRNAs, 2 rRNAs, a non-coding region, and a control region (D-loop). The phylogenetic analysis using Bayesian Inference validated the taxonomic status of P. granulosus, showing the close relationship with the other two species from the genus Pachytriton.
There are currently only ten species of Pachytriton (Caudata: Salamandridae) distributed in southeastern China (Frost Citation2020). The mitochondrial genome (mitogenome), as an important molecular marker, has been utilized for phylogenetic analysis. However, only two complete mitogenomes of Pachytriton species could be obtained from Genbank database at present (Fang et al. Citation2017; Hu et al. Citation2020). The Bingzhi's stout newt (Pachytriton granulosus Chang, 1933) is a Chinese endemic newt, which mainly inhabits mountainous areas of Zhejiang, China (Frost Citation2020). Here, we sequenced and annotated the complete mitogenome of P. granulosus, representing the third mitochondrial genome from the genus Pachytriton.
The specimen of P. granulosus was collected at Jinzijian (N29.797983°, E118.963999°) from Chun’an County, Zhejiang Province, China. The collected specimen was stored in 90% ethanol and deposited at the Museum of Laboratory of Amphibian Diversity Investigation (contact person: Guo-Hua Ding, E-mail: [email protected]) at Lishui University under the voucher number LSU20200827JZJ01. Whole genomic DNA was extracted from muscle tissue using the EasyPure genomic DNA kit (TransGen Biotech Co, Beijing, China). The library of WGS (21.84 G) was carried out on an Illumina NovaSeq 6000 platform (Novogene Bioinformatics Technology Co. Ltd., Tianjin, China) for paired-end sequencing of 150 bp. The mitogenome as a byproduct of whole genome sequencing was assembled using NOVO Plasty 3.7 (Dierckxsens et al. Citation2017), and annotated by MITOS Web Server (Matthias et al. Citation2013).
We obtained the complete mitogenome of P. granulosus of 16,293 bp (Genbank accession: MW056200). This mitogenome encoded 13 protein-coding genes (PCGs), 22 tRNAs, 2 rRNAs (16S and 12S), a non-coding region (NCR) of an L-strand replication origin, and a control region (D-loop). The overall nucleotide composition was 33.56% of A, 28.52% of T, 14.68% of G, and 23.24% of C, demonstrating a bias of higher AT content (62.08%) than GC content (37.92%). The gene arrangement pattern and transcription directions were identical to previous studies in the genus Pachytriton (Fang et al. Citation2017; Hu et al. Citation2020). Among the 13 PCGs, the ATP8 was the shortest (168 bp), while the ND5 was the longest (1,812 bp). All PCGs initiated with ATG as a start codon, except for COI and ND1 genes, which began with GTG. Nine PCGs (ND1, ND2, COI, ATP8, ATP6, ND3, ND4L, ND5, and ND6) end with complete stop codons (AGA, TAA, and TAG), and the other four PCGs end with T (COII and COIII genes) or TA (CYTB gene) as the incomplete stop codons, which were presumably completed as TAA by post-transcriptional polyadenylation (Anderson et al. Citation1981). All genes were encoded on the heavy strand, except eight tRNAs (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer,tRNAGlu, and tRNAPro) and ND6 gene, which were encoded on the light strand. The D-loop (733 bp) was found between tRNAPro and tRNAPhe, and the NCR (30 bp) is found between tRNAAsn and tRNACys.
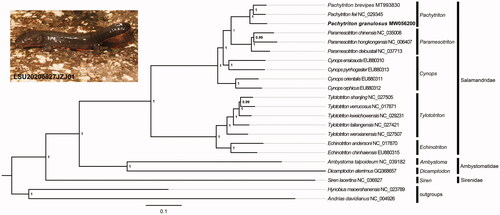
To validate the phylogenetic position of P. granulosus, a Bayesian Inference (BI) tree was constructed on MrBayes v3.2.2 using the 13 PCGs (1,1400 bp) in mitogenomes of 22 species. Hynobius maoershanensis (Caudata: Hynobiidae) and Andrias davidianus (Caudata: Cryptobranchidae) were chosen as the outgroups. We obtained the best-fit substitution model (GTR + I + G) by MrModelTest 2.3 (Nylander Citation2004) in PAUP 4.0. Four parallel runs of Markov Chain Monte Carlo (MCMC) were analyzed for 1,000,000 generations, sampling every 1000 generations and discarded 1000 trees as burn-in. The three Pachytriton species showed a close phylogenetic relationship with other species in the family Salamandridae, which clustered in a monophyletic group (). The phylogenetic analysis result was consistent with the previous research (Hu et al. Citation2020). The p-distances between P. granulosus and other two Pachytriton species were both more than 7.0% based on the 13 PCGs using MEGA 5.05. The new complete mitogenomes of P. brevipes in this study will contribute to the further research of species identification and molecular evolution of the genus Pachytriton.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The mitogenome data supporting this study are openly available in GenBank at [https://www.ncbi.nlm.nih.gov/nuccore/MW056200]. Reference number [Accession number: MW056200]. SRA and BioSample accession numbers are [https://www.ncbi.nlm.nih.gov/sra/SRR12746564], [https://www.ncbi.nlm.nih.gov/biosample/SAMN16294863], respectively.
Additional information
Funding
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290(5806):457–465.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Fang K, Zhang CL, Guo WB, Lifu Qian LF, Wu J, Zhang BW. 2017. Mitochondrial genome of Pachytriton feii (Urodela: Salamandridae). Mitochondrial DNA A DNA Mapp Seq Anal. 28(2):288–289.
- Frost DR. 2020. Amphibian species of the world: an online reference, version 6.0. New York (NY): American Museum of Natural History. [accessed 2020 Oct 2]. http://research.amnh.org/herpetology/amphibia/index.php/.
- Hu HL, Zhong JJ, Ma L, Lin ZH, Ding GH. 2020. Complete mitochondrial genome of Black-spotted Stout Newt (Pachytriton brevipes) and its phylogenetics analysis. Mitochondrial DNA B. doi: 10.1080/23802359.2020.1847619.
- Matthias B, Alexander D, Frank J, Fabian E, Catherine F, Guido F, Joern P, Martin M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.