Abstract
We report the first complete mitochondrial genome of an important pest of timber, the drywood termite Cryptotermes havilandi. The gene content and synteny of the mitochondrial genome of C. havilandi is identical to that of other termite species reported to date. It is composed 13 protein-coding genes, two ribosomal RNA genes, and 22 transfer RNA genes. Our phylogenetic tree, that includes the mitochondrial genomes of 14 species of Kalotermitidae, including C. havilandi, resolves the phylogenetic position of C. havilandi within Kalotermitidae.
Main text
Cryptotermes havilandi Sjöstedt, 1900 (Isoptera: Kalotermitidae) is an important pest of structural lumber and sheltered wood (Su and Scheffrahn Citation2000). Although it is now distributed across the tropical and subtropical regions, C. havilandi originated from Africa, and has been introduced outside its native range largely by the intermediary of human transportation (Evans Citation2011; Evans et al. Citation2013). It is now invasive in various Caribbean islands, Guiana, Surinam, Brazil, Madagascar, the Comores, and India (Evans et al. Citation2013). Despite its economic importance, the mitochondrial genome of C. havilandi has not been sequenced yet. Here, we provide the first complete mitochondrial genome sequence of a C. havilandi extracted from the sample CAM101 collected on 7th of April 2015 in an abandoned wooden house in northern Cameroon, Africa (N04°42′25″ E009°43′08″), by the authors.
We sequenced C. havilandi (GenBank: MW208858) mitochondrial genome using Illumina HiSeq2000. The genome was assembled using the clc suite of programs as described by Bourguignon et al. (Citation2015). The total length of the complete mitochondrial genome of C. havilandi is 15,559bp. As in other mitochondrial genomes of termites previously sequenced (Cameron and Whiting Citation2007; Cameron et al. Citation2012; Bourguignon et al. Citation2015, Citation2016, Citation2017; Wu et al. Citation2018; Wang et al. Citation2019), the mitochondrial genome of C. havilandi is composed of 13 protein-coding genes (following the order: nad2, cox1, cox2, atp8, atp6, cox3, nad3, nad5, nad4, nad4l, nad6, cytb, and nad1), two ribosomal RNA genes (rnl and rns) and 22 transfer RNA genes (following the order: Ile, Gln, Met, Trp, Cys, Tyr, Leu(UUR), Lys, Asp, Gly, Ala, Arg, Asn, Ser(AGN), Glu, Phe, His, Thr, Pro, Ser(UCN), Leu(CUN), and Val). The GC-content is 34%. Our results confirm that termite mitochondrial genomes are stable in gene content and preserved their synteny.
To shed light on the phylogenetic position of C. havilandi within the Kalotermitidae, we reconstructed a phylogenetic tree that included all mitochondrial genomes of Kalotermitidae sequenced to date, including the mitochondrial genome of C. havilandi, and three outgroups: Zootermopsis angusticolis (Isoptera: Archotermopsidae), Porotermes adamsoni (Isoptera: Termopsidae) and Coptotermes sepangensis (Isoptera: Rhinotermitidae) (). All genes were aligned separately using MAFFT v. 7.3 (Katoh and Standley Citation2013), concatenated, and the phylogenetic tree was reconstructed using MrBayes v. 3.2.1 (Ronquist et al. Citation2012). The parameters of the phylogenetic analysis were set as described by Bourguignon et al. (Citation2017). Overall, our phylogenetic tree confirms the monophyly of Cryptotermes, within which C. havilandi is nested.
Figure 1. Bayesian phylogenetic tree of all species of Kalotermitidae sequenced to date. Numbers in nodes state for posterior probabilities and the scale indicates 6% genetic variation for its length. GenBank accession numbers are given in brackets. “*” mark the studied species.
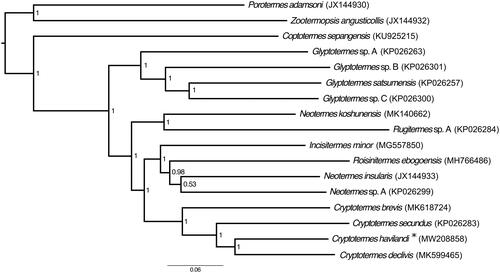
The genus Cryptotermes includes several invasive species that cause major economic losses in the world (Evans et al. Citation2013). Surprisingly, very few studies have used molecular markers to study the population genetics of Cryptotermes species. In this paper, we provide the mitochondrial genome of one of the most important termite pest. The new mitochondrial genome presented here will help to understand how the major termite pests have been introduced around the world.
Author contribution
PS and JS conceived the study, carried out bioinformatics analyses and wrote the paper. PA facilitated the fieldwork in Cameroon.
Acknowledgment
The authors thank Thomas Bourguignon for his valuable insights. We are grateful to Raphael Onana and to the people of village Ebogo II for their hospitality and help in biological material collection.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW208858. The associated SRA, BioProject, and Bio-Sample numbers are SRR13287752, PRJNA687161 and SAMN17119085, respectively.
The sample CAM101 is available in termite collection at Czech University of Life Sciences, Prague, Czech Republic in both states, as an 80% ethanol voucher sample and in RNAlater preservative (contact: [email protected]).
Additional information
Funding
References
- Bourguignon T, Lo N, Cameron SL, Šobotník J, Hayashi Y, Shigenobu S, Watanabe D, Roisin Y, Miura T, Evans TA. 2015. The evolutionary history of termites as inferred from 66 mitochondrial genomes. Mol Biol Evol. 32(2):406–421.
- Bourguignon T, Lo N, Šobotník J, Ho SYW, Iqbal N, Coissac E, Lee M, Jendryka MM, Sillam-Dussès D, Křížková B, et al. 2017. Mitochondrial phylogenomics resolves the global spread of higher termites, ecosystem engineers of the tropics. Mol Biol Evol. 34(3):589–597.
- Bourguignon T, Lo N, ŠobotnÍk J, Sillam-Dussès D, Roisin Y, Evans TA. 2016. Oceanic dispersal, vicariance and human introduction shaped the modern distribution of the termites Reticulitermes, Heterotermes and Coptotermes. Proc Biol Sci. 283(1827):20160179.
- Cameron SL, Lo N, Bourguignon T, Svenson GJ, Evans TA. 2012. A mitochondrial genome phylogeny of termites (Blattodea: Termitoidae): robust support for interfamilial relationships and molecular synapomorphies define major clades. Mol Phylogenet Evol. 65(1):163–173.
- Cameron SL, Whiting MF. 2007. Mitochondrial genomic comparisons of the subterranean termites from the genus Reticulitermes (Insecta: Isoptera: Rhinotermitidae). Genome. 50(2):188–202.
- Evans TA. 2011. Invasive termites. In: Bignell DE, Roisin Y, Lo N, editors. Biology of termites: a modern synthesis. Dordrecht; Heidelberg; London; New York: Springer; p. 519–562.
- Evans TA, Forschler BT, Grace JK. 2013. Biology of invasive termites: a worldwide review. Annu Rev Entomol. 58:455–474.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Su NY, Scheffrahn R. 2000. Termites as pests of buildings. In: Abe T, Bignell DE, Higashi M, editors. Termites: evolution, sociality, symbioses, Ecology. Dordrecht, The Netherlands: Kluwer Academic Publishers; p. 437–453.
- Wang M, Buček A, Šobotník J, Sillam-Dussès D, Evans TA, Roisin Y, Lo N, Bourguignon T. 2019. Historical biogeography of the termite clade Rhinotermitinae (Blattodea: Isoptera). Mol Phylogenet Evol. 132(1):100–104.
- Wu LW, Bourguignon T, Šobotník J, Wen P, Liang WR, Li HF. 2018. Phylogenetic position of the enigmatic termite family Stylotermitidae (Insecta: Blattodea). Invert Systematics. 32(5):1111–1117.
