Abstract
Olenecamptus bilobus Fabricius is widely distributed in some parts of Southeast and East Asia whose larvae bore under the bark of at least eleven plant families. The complete mitochondria genome of O. bilobus was 15,262 bp in length, with 37 genes, including 12 protein-coding genes (PCGs), 23 tRNA genes (tRNAs), and 2 rRNA genes (rRNAs). The A + T content is 76.91%, showing strong AT skew. Phylogenetic analysis indicated that O. bilobus had a close relationship with Olenecamptus subobliteratus Pic.
Introduction
Olenecamptus bilobus is a common pest whose larvae bore under the bark of at least eleven plant families such as mulberry, willow, poplar, oyster, oak, sassafras, tussah, maple, banyan, claw beetle, pineapple etc. (Chen et al. Citation1959). According to the few reports in the past, we can know that O. bilobus widely distributed in some parts of Southeast and East Asia (Hua et al. Citation2009; Saha et al. Citation2013). Olenacamptus taiwanus Dillon & Dillon was once mistaken as a subspecies of this species which reflected the necessity of mitochondrial genome for species identification (Wang and Tang Citation2018 Wang et al. Citation2019; Su and Wang Citation2020; Zhou et al. Citation2020). Therefore, it is necessary to study the complete mitochondrial DNA (mtDNA) genome of O. bilobus which provides a theoretical basis for future research on Olenecamptus chevrolat.
In this study, specimens of O. bilobus were collected from the Qingxiu Mountain (22°47’N, 108°23’E) of Nanning City (Guangxi Autonomous Region, China) on a banyan tree. The total genomic DNA was extracted following the modified CTAB DNA extraction protocol and stored at Guangxi Key Laboratory of Agric-Environment and Agric-Products Safety (The city of Nanning, China) with sample number of SZHT0606G148. Then library was constructed and pair-end was sequenced (2*150 bp) with HiSeq (Illumina, San Diego, CA). Approximately 11.10 G of raw data and 11.00 G of clean data were obtained for sequence assembly by SPAdes (version 3.9) (Bankevich et al. Citation2012). MITOS (http://mitos.bioinf.uni-leipzig.de/index.py) was used for mitochondrial genome annotation (Bernt et al. Citation2013).
The complete mitochondrial genome of O. bilobus is a closed circular molecule 15,262 bp in length (GenBank accession number MT740324) and constitutive of 37 genes. These genes contain 12 protein-coding genes (PCGs), 23 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and 1 A + T region (D-loop). The single A + T region is 613 bp in length. The nucleotide composition of the O. bilobus mitogenome was A (38.36%), T (38.55%), G (9.24%), C (13.85%). The A + T content is 76.91%, showing strong AT skew.
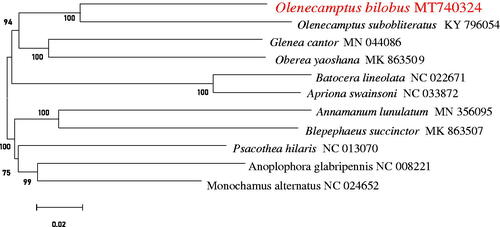
Molecular Evolutionary Genetics Analysis Version 7.0 (MEGA 7.0) was used to make phylogenetic analysis among Lamiinae species by Neighbor-Joining method with 1000 bootstrap replicates (Kumar et al. Citation2016). The results showed that mtDNA of O. bilobus had a close relationship with that of O. subobliteratus ().
Acknowledgements
We appreciated the sequencing service provided by Huitong biotechnology Co. Ltd (Shenzhen, China).
Disclosure statement
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at: https://www.ncbi.nlm.nih.gov/nuccore/MT740324. Associated BioProject, SRA, and BioSample accession numbers are https://www.ncbi.nlm.nih.gov/bioproject/PRJNA663376, https://www.ncbi.nlm.nih.gov/sra/SRX9145194, and SAMN16131500, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler FP. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chen SX, Xie YZ, Deng GF. 1959. Economic insect fauna of China (Volume I, Coleoptera). Vol. 1. Beijing: Science Press; p. 96.
- Hua LZ, Nara H, Samuelson GA, Lingafelter SW. 2009. Iconography of Chinese longicorn beetles (1406 species) in color. Guangzhou: Sun Yat-sen University Press; p. 381.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Saha S, Özdikmen H, Biswas MK, Raychaudhuri D. 2013. Exploring flat faced longhorn beetles (Cerambycidae: Lamiinae) from the reserve forests of Dooars, West Bengal, India. ISRN Entomol. 2013:1–8.
- Su RR, Wang XY. 2020. The complete mitochondrial genome of the Megopis sinica White (Coleoptera: Cerambycidae: Prioninae). J Mitochondrial DNA Part B Res. 5(1):236–237.
- Wang Q, Tang G. 2018. The mitochondrial genomes of two walnut pests, Gastrolina depressa depressa and G. depressa thoracica (Coleoptera: Chrysomelidae), and phylogenetic analyses. PeerJ. 6:e4919.
- Wang XY, Zheng XL, Lu W. 2019. The complete mitochondrial genome of an Asian longicorn beetle Glenea cantor (Coleoptera: Cerambycidae: Lamiinae). Mitochondrial DNA B Resour. 4(2):2906–2907.
- Zhou SC, Zheng XL, Lu W. 2020. The complete mitochondrial genome of an Asian longicorn beetle Dorysthenes granulosus (Coleoptera: Cerambycidae: Prioninae). Mitochondrial DNA B Resour. 5(1):673–674.