Abstract
Cacopsylla citrisuga (Yang & Li) is an important pest-threatening Citrus and Poncirus plants (Rutaceae) and a newly identified insect vector of citrus Huanglongbing. The complete mitochondrial genome of C. citrisuga was 14,906 bp in length, with 37 genes, including 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs). The phylogenetic trees inferred from Bayesian inference and maximum likelihood analyses confirmed C. citrisuga as a member of the genus Cacopsylla. Our phylogenetic analyses suggested that the Cacopsylla is paraphyletic, and confirmed C. citrisuga as a member of clade-I under Cacopsylla. The complete mitochondrial genome of C. citrisuga will provide important information for the phylogeny and evolution analysis of Cacopsylla.
Huanglongbing (HLB), known as citrus greening disease, is associated with phloem-restricted alpha-proteobacterium: ‘Candidatus Liberibacter asiaticus,’ ‘Ca. L. africanus,’ and ‘Ca. L. americanus’ (Bové Citation2006). Cacopsylla citrisuga (Yang & Li) (Hemiptera: Psyllidae), is an important pest of Citrus and Poncirus plants (Rutaceae), has been proved to be a new insect vector of ‘Candidatus Liberibacter asiaticus’ (Cen et al. Citation2012). This pest belongs to the genus Cacopsylla Ossiannilsson, 1970, which is the largest psyllid genus comprising ca. 470 species (https://hemiptera-databases.org/). To date, 359 complete or near complete mitogenomes from species of the psyllids (family Psylloidea) have been sequenced, among which only 24 are from the genus Cacopsylla; in addition, the evolution relationship of Cacopsylla citrisuga with other psyllids is still less described. In this study, we analyzed and reported the first mitogenomes of Cacopsylla citrisuga and built its phylogenetic history with other psyllids.
The adult specimens used in this study were collected at Mengxiu, Yunnan, China (24.075618°N, 97.741441°E) in June 2018. The collected specimen was kept in 80% ethyl alcohol and deposited in Insect Collection of Zhejiang A&F University (ZAFU-INSECT-Y501). Whole genome DNA was extracted using the QIAamp DNA Micro Kit (Qiagen Hilden, Germany) and sequenced on a HiSeq 2000 platform (Illumina, USA) with a paired-end 150-bp sequencing strategy. Clean data screening and de novo assembly also follows the method of Wang et al. (Citation2018). We obtained 14,906 base-pair of assembled, circular mitochondrial genomic sequences for C. citrisuga (GenBank accession number: MT990978). The assembled psyllid mitochondrial genome fit well within the conserved nature of the mitochondrial genomes of Hemiptera (Wang et al. Citation2015): contain 13 protein-encoding genes (PCGs; COI-III, ND1-6, ND4L, ATP6, ATP8, and CytB), 22 tRNA genes, two ribosomal RNAs and a non-coding A + T-rich region (Supplementary Table S1; Figure S1). The overall base composition of the mitochondrial genome is A (38.2%), T (34.5%), C (17.7%), G (9.6%).
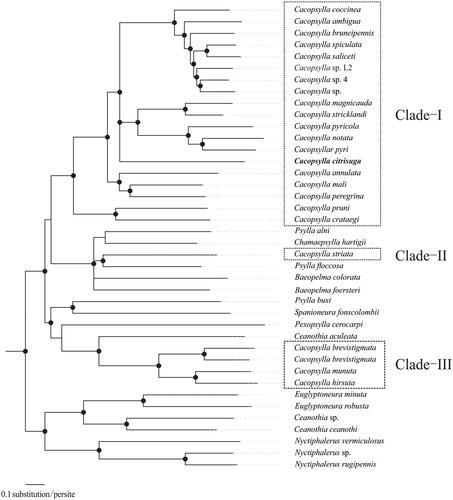
Phylogenetic trees were constructed based on 13 PCGs sequences from 40 species (Supplementary Table S2) by Bayesian Inference (BI) and maximum likelihood (ML) analyses. The 40 mitochondrial sequences were aligned by the MAFFT version 7 software (Katoh and Standley Citation2013). Bayesian Inference was conducted by MrBayes 3.2.1 (Ronquist and Huelsenbeck Citation2003) and maximum likelihood (ML) analyses implemented in IQ-TREE 1.5.5 (Nguyen et al. Citation2015). Cacopsylla was recovered as paraphyletic clades in the present study, consistently with previous paraphyletic research (Percy et al. Citation2018; Cho et al. Citation2019). Both BI and ML analyses strongly supported C. citrisuga as a member of Cacopsylla, and C. citrisuga was embraced in a clade consisting C. coccinea, C. ambigua, C. bruneipennis, C. spiculata, C. saliceti, C. sp. L2, C. sp. 4, C. sp., C. magnicauda, C. stricklandi, C. pyricola, C. notata, C. pyri, with strong supports (PP ≥ 0.90; BP ≥ 95; ). The availability of this additional genetic information promises to open the way for studies of phylogeny of Cacopsylla. This report is the first molecular characterization of complete mitogenome of the important agricultural pest, C. citrisuga. The availability of this additional genetic information will be useful for further investigating the evolutionary relationships within Cacopsylla.
Figure 1. Bayesian phylogenetic inferred from 40 psyllids mitogenome sequences based on 13 PCGs. The node support values indicate the Bayesian posterior probabilities (PP) and the bootstrap (BP) values. Cycles on the nodes indicated BI posterior probabilities ≥ 0.90 and ML bootstrap values ≥ 95 for clades.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT990978
Additional information
Funding
References
- Bové JM. 2006. Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J Plant Pathol. 88:7–37.
- Cen Y, Zhang L, Xia Y, Guo J, Deng X, Zhou W, Sequeira R, Gao J, Wang Z, Yue J, et al. 2012. Detection of ‘Candidatus Liberibacter Asiaticus’ asiaticus’ in Cacopsylla (Psylla) citrisuga (Hemiptera: Psyllidae). Florida Entomologist. 95(2):304–311.
- Cho G, Malenovský I, Lee S. 2019. Higher-level molecular phylogeny of jumping plant lice (Hemiptera: Sternorrhyncha: Psylloidea). Syst Entomol. 44(3):638–651.
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Percy DM, Crampton-Platt A, Sveinsson S, Lemmon AR, Lemmon EM, Ouvrard D, Burckhardt D. 2018. Resolving the psyllid tree of life: phylogenomic analyses of the superfamily Psylloidea (Hemiptera). Syst Entomol. 43(4):762–776.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Wang Y, Chen J, Jiang LY, Qiao GX. 2015. Hemipteran mitochondrial genomes: features, structures and implications for phylogeny. Int J Mol Sci. 16(6):12382–12404.
- Wang Y, Lu J, Beattie GA, Islam MR, Om N, Dao HT, Van Nguyen L, Zaka SM, Guo J, Tian M, et al. 2018. Phylogeography of Diaphorina citri (Hemiptera: Liviidae) and its primary endosymbiont, ‘Candidatus Carsonella ruddii’: an evolutionary approach to host–endosymbiont interaction. Pest Manag Sci. 74(9):2185–2194.
