Abstract
Bar-tailed Treecreeper Certhia himalayana usually lives in coniferous or mixed broadleaf-conifer forests, often crawling along the trunk. In this study, we first sequenced and described the complete mitochondrial genome and phylogeny of C. himalayana. The whole genome of C. himalayana was 16,852 bp in length, and contained 13 protein-coding genes, 22 transfer RNA genes, 2 ribosome RNA genes, and 1 non-coding control regions. The overall base composition of the mitochondrial DNA was 25.1% for A, 29.2% for T, 14.5% for C, 31.2% for G, with a GC content of 45.7%. A phylogenetic tree strongly supported that C. himalayana closely related with Family Troglodytidae by highly probability.
Bar-tailed Treecreeper (Certhia Himalayana Vigors, 1832) inhabits coniferous forest or mixed broadleaf-conifer forest at an altitude of 1000-3500 meters, occasionally occurs in the shrub to c.500 m (BirdLife International Citation2016). C. himalayana occurs in Central Asia and southwest of China (Yunnan, Sichuan and Shanxi provinces) (Harrap and Quinn Citation1996). This species has an extremely large range, and hence does not approach the thresholds for Vulnerable under the range size criterion (BirdLife International Citation2016). Molecular studies supported C. himalayana closely related with Sittidae and Troglodytidae by highly probability based on mitochondrial genes, nuclear genes, both nuclear and mitochondrial genes, or mitochondrial and morphological data (Jonsson and Fjeldsa Citation2006). Barker et al (Citation2002) and Fregin et al (Citation2012) also proved it used nucleotide sequence or mitochondrial and nuclear genes. The complete mitochondrial genome of C. himalayana has not been determined and characterized until now. Therefore, the aim of this study was to first assemble and characterize the complete mitochondrial genome of C. himalayana.
The specimen (Duan-025) was collected from Zixi Mountain (25.02 N, 101.41E), which was located central Yunnan Province in China, and stored at the Herbarium of Southwest Forestry University. The total mitochondrial DNA was extracted from the muscle tissue using the Ezup Column Animal Genomic DNA Kit (Sangon, Shanghai, China). The mitogenome was sequenced using Illumina Hiseq sequencing platform (Illumina, San Diego, CA) and assembled with A5-miseq v20150522 (Coil et al. Citation2015). All library constructions and sequencing were performed at Sangon Bio Co., Ltd., Shanghai, China. The complete mitochondrial genome was annotated using MITOS Web server (http://mitos2.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013) and OGDRAW 1.3.1 (Greiner et al. Citation2019). The complete mitochondrial genome of C. himalayana was submitted to the NCBI database under the accession number MN624167. A maximum likelihood (ML) tree was implemented in RAxML v7.7.1 (Stamatakis Citation2014) under the GTR-Gamma model, and node support was calculated with 1000 bootstrap replications (Wei et al. Citation2016).Sequences of Lorius chlorocercus and Psittacula alexandr obtained from GenBank (MN_515396 and MK_986660) were used as outgroups to root trees following Jonsson and Fjeldsa (Citation2006).
The complete mitochondrial genome of C. himalayana was found to be a circular double-stranded 16,814 bp in length. A total of 38 mitochondrial genes were identified, including 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and 1 non-coding control region (D-loop). All the 13 protein coding genes contained the same start condon TTA/TAA, except that nad6 and nad4 gene started with ATG, nad5 gene started with TCT, cox1 gene started with CCT. Furthermore, eleven of the PCGs used complete ATT/ATG or incomplete A(TA) stop codon, and other two types were AGG[nad6], ATC[atp8].
Among these genes, nad6 and 8 tRNAs (trnE, trnP, trnS, trnY, trnC, trnN, trnA and trnQ) were located on the light strand (L-strand), while all of the remaining genes were located on the heavy strand (H-strand). The overall base composition of C. himalayana mitogenome was 25.1% for A, 29.2% for T, 14.5% for C, 31.2% for G, A + T content is 54.3%, which is higher than G + C content of 45.7%, similar to other Psittaciformes (Barker Citation2014; Song et al. Citation2018).
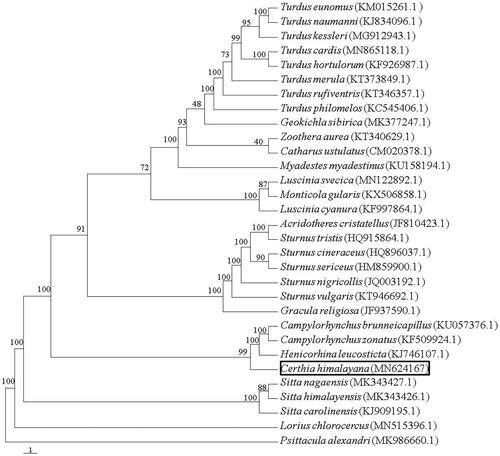
The reconstructed phylogenetic tree showed that C. himalayana grouped with Family Troglodytidae (Campylorhynchus brunneicapillus, C. zonatus and Henicorhina leucosticta) with strong support (100% bootstrap support value) by the analyses of protein-coding genes (). C. himalayana closely related with Family Troglodytidae, which is congruent with previous studies (Jonsson and Fjeldsa Citation2006). The complete mitochondrial genome of C. himalayana reported here will be useful for future population genetic studies of this species and will provide essential genome resources for the ecologically important species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number [MN624167], or available from the corresponding author.
Additional information
Funding
References
- Barker KF. 2014. Mitogenomic data resolve basal relationships among passeriform and passeridan birds. Mol Phylogenetics Evol. 79:313–324.
- Barker FK, Barrowclough GF, Groth JG. 2002. A phylogenetic hypothesis for passerine birds: taxonomic and biogeographic implications of an analysis of nuclear dna sequence data. Proc Biol Sci. 269(1488):295–308.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- BirdLife International 2016. Species factsheet: Certhia himalayana. [Downloaded on 30 Aug 2020] http://www.birdlife.org.
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589.
- Fregin S, Haase M, Olsson U, Alström P. 2012. New insights into family relationships within the avian superfamily Sylvioidea (Passeriformes) based on seven molecular markers. BMC Evol Biol. 12:157.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Harrap S, Quinn D. 1996. Tits, nuthatches and treecreepers. London: A&C Black.
- Jonsson KA, Fjeldsa J. 2006. A phylogenetic supertree of oscine passerine birds (aves: passeri). Zool Scripta. 35(2):149–186.
- Song S, Qin J, Luo J, Li D, Jiang B, Chang C. 2018. Analysis of complete mitochondrial genome sequence of kessleri thrush, turdus kessleri (passeriformes, turdidae). Mitochondrial Dna Part B. 3(2):818–819.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wei M, Liu Y, Guo H, Zhao F, Chen S. 2016. Characterization of the complete mitochondrial genome of Cynoglossus gracilis and a comparative analysis with other Cynoglossinae fishes. Gene. 591(2):369–375.