Abstract
Adonis amurensis Regel et Radde is an important cardiac folk medicinal plant which endemic to Northeast Asia. We determined the first complete chloroplast genome of A. amurensis using genome skimming approach. The cp genome was 157,032 bp long, with a large single-copy region (LSC) of 86,218 bp and a small single-copy region (SSC) of 18,212 bp separated by a pair of inverted repeats (IRs) of 26,301 bp. It encodes 129 genes, including 84 protein-coding genes, 37 tRNA genes, and 8 ribosomal RNA genes. We also reconstructed the phylogeny of Adonideae and Isopyreae using maximum likelihood (ML) method, including our data and previously reported cp genomes of related taxa. The phylogenetic analysis indicated that A. amurensis is close related with Adonis sutchuenensis.
The genus Adonis L. (Ranunculaceae), native to Europe and Asia, comprises 32 annual or perennial herbaceous species. Due to their cardiac-enhancing effects, Adonis spp. have long been used in European and Chinese folk medicine (Shang et al. Citation2019). Adonis amurensis Regel et Radde is morphologically distinguished from other taxa of Adonis by unbranched glabrous stem, long leaf petioles branched two or three times, one flower with eight to nine sepals, and sepals longer to that of petals (Gorovoy and Gurzenkov Citation1969; Wang Citation1980; Nishikawa and Kadota Citation2006; Son Citation2015). However, because of the morphological characters that are shared among objective distinctive characteristics of the taxa within the genus and the diverse morphological variation of A. amurensis, it has been difficult to identify taxa and investigate their phylogenetic relationships, and hence taxonomic status of A. amurensis has been treated in very contrasting ways (Son Citation2015; Son et al. Citation2018). Despite recent studies on the genus Adonis from East Asia (Kaneko et al. Citation2008; Son et al. Citation2016; Son et al. Citation2017), the taxonomic and phylogenetic relationships among taxa still need to be confirmed by more molecular evidences. By taking advantages of next-generation sequencing technologies that efficiently provide the chloroplast (cp) genomic resources of our interested species, we can rapidly access the abundant genetic information for phylogenetic research and conservation genetics (Li et al. Citation2017; Liu et al. Citation2017). Therefore, we sequenced the whole chloroplast genome of A. amurensis to elucidate its phylogenetic relationship within Ranunculaceae.
Total genomic DNA was extracted from silica-dried leaves collected from the campus of Tonghua Normal University using a modified CTAB method (Doyle and Doyle Citation1987). The voucher specimen (sfxyyyxycjzh20200429) was collected and deposited in the Herbarium of Tonghua Normal University (41°44′40.69″, 125°58′57.63″, 427.3). DNA libraries preparation and pair-end reads sequencing were performed on the Illumina NovaSeq 6000 platform. The cp genome was assembled via NOVOPlasty (Dierckxsens et al. Citation2017), using the A. sutchuenensis cp genome (MK569470, Zhai et al. Citation2019) as a reference. Gene annotation was performed via the online program Dual Organellar Genome Annotator (DOGMA; Wyman et al. Citation2004). Geneious R11 (Biomatters Ltd., Auckland, New Zealand) was used for inspecting the cp genome structure.
The complete cp genome of A. amurensis (GenBank accession MW042677) was 157,032 bp long consisting of a pair of inverted repeat regions (IRs with 26,301 bp) divided by two single-copy regions (LSC with 86,218 bp; SSC with 18,212 bp). The overall GC contents of the total length, LSC, SSC, and IR regions were 38.0%, 36.2%, 31.5% and 43.1%, respectively. The genome contained a total of 129 genes, including 84 protein-coding genes, 37 tRNA genes and 8 rRNA genes.
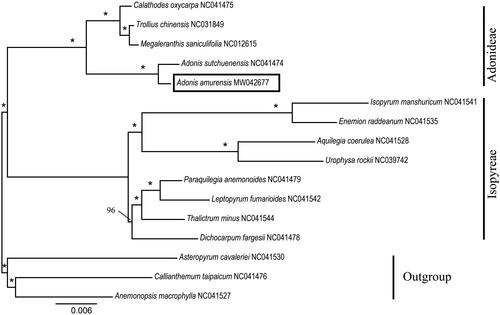
We used a total of 15 additional complete cp genomes of the Ranunculaceae species to clarify the phylogenetic position of A. amurensis. Asteropyrum cavaleriei (NC041530), Callianthemum taipaicum (NC041476) and Anemonopsis macrophylla (NC041527) were used as the outgroup. We reconstructed a phylogeny employing the GTR + G model and 1000 bootstrap replicates under the maximum-likelihood (ML) inference in RAxML-HPC v.8.2.10 on the CIPRES cluster (Miller et al. Citation2010). The ML tree () was consistent with the most recent phylogenetic study on Ranunculaceae (Son et al. Citation2016; Zhai et al. Citation2019). A. amurensis exhibited the closest relationship with Adonis sutchuenensis.
Disclosure statement
The authors are grateful to the opened raw genome data from public database. The authors report no conflicts of interest and are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW042677.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Gorovoy PG, Gurzenkov NN. 1969. Adonis ramosa Franch. (Ranunculaceae), a new species for flora of the USSR and some critical remarks on the Far Eastern species of Adonis L. Botanicheskii Zhurnal. 54:139–e143.
- Kaneko S, Nakagoshi N, Isagi Y. 2008. Origin of the endangered tetraploid Adonis ramosa (Ranunculaceae) assessed with chloroplast and nuclear DNA sequence data. Acta Phytotaxonomica et Geobotanica. 59(2):165e174.
- Li P, Lu RS, Xu WQ, Ohi-Toma T, Cai MQ, Qiu YX, Cameron KM, Fu CX. 2017. Comparative Genomics and Phylogenomics of East Asian Tulips (Amana, Liliaceae). Front Plant Sci. 8:451.
- Liu LX, Li R, Worth JRP, Li X, Li P, Cameron KM, Fu CX. 2017. The complete chloroplast genome of Chinese bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of Fagales. Front Plant Sci. 8:968.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Comp Environ Workshop. 14:1–8.
- Nishikawa T, Kadota Y. 2006. Adonis. In: Iwatsuki K, Boufford DE, Ohba H, editors. Flora of Japan, vol. IIa. Tokyo, Japan: Kodansha Ltd.
- Shang X, Miao X, Yang F, Wang C, Li B, Wang W, Pan H, Guo X, Zhang Y, Zhang J. 2019. The genus Adonis as an important cardiac folk medicine: a review of the ethnobotany, phytochemistry and pharmacology. Front Pharmacol. 10:25.
- Son DC, Lee J, Eo KS, Park BK, Choi K. 2018. Adonis amurensis var. pilosissima var. nov. a new variety of Adonis amurensis (Ranunculaceae) from east asia. J Asia Pacific Biodiversity 11 (1):49–55.
- Son DC, Park BK, Chang KS, Choi K, Shin CH. 2017. Cladistic analysis of the section Adonanthe under genus Adonis L. (Ranunculaceae) from East Asia. J Asia-Pac Biodivers. 10(2):232e236–232e236.
- Son DC, Park BK, Ko SC. 2016. Phylogenetic study of the section Adonanthe of genus Adonis L. (Ranunculaceae) based on ITS sequences. Korean Journal of Plant Taxonomy. 46(1):1e12–1e12.
- Son DC. 2015. A systematic study on the section Adonanthe of genus Adonis L. (Ranunculaceae) in East Asia. PhD thesis. Daejeon, Korea: Hannam University.
- Wang WT. 1980. Adonis. In: Wang WT, editor. Flora of reipublicae popularis sinicae, vol. 28. Beijing: Agendae Academiae Sinicae Edita. p. 246e255. in Chinese).
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zhai W, Duan X, Zhang R, Guo C, Li L, Xu G, Shan H, Kong H, Ren Y. 2019. Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the Ranunculaceae. Mol Phylogenet Evol. 135:12–21.