Abstract
The Gymnobelideus leadbeateri (Leadbeater’s Possum) is listed as Critical Endangered on the International Union for Conservation of Nature (IUCN) Red List. We assembled the complete mitochondrial genome for the G. leadbeateri and characterized it to provide informative data for forthcoming studies for understanding its evolution and conservation genetics. The G. leadbeateri mitogenome is 16,812 bp long and encodes 13 protein-coding genes (PCGs), two ribosomal RNA genes (12S rRNA and 16S rRNA) and 22 transfer RNA (tRNA) genes. Phylogenetic analysis based on complete mitogenome shows that G. leadbeateri is related to Petaurus breviceps (sugar glider) and Dactylopsila trivirgata (striped possum).
The Gymnobelideus leadbeateri McCoy is distributed only in Victoria state in southeast Australia (Harley Citation2004). Its population is estimated to be 1,100 ∼ 11,000 and the species mainly lives in old trees’ hollows (Woinarski and Burbidge Citation2016). Gymnobelideus leadbeateri has been listed on Critically Endangered IUCN Red List with a declining population across its range (Woinarski and Burbidge Citation2016), whereas its habitat is threatened by forest fire and timber industry (Lindenmayer et al. Citation1991). In present study, we assembled the mitochondrial genome of G. leadbeateri to know more about breeding and genetics of this species and protect it better.
Sample collected from a female Leadbeater’s possum at Yellingbo Nature Conservation Reserve, Australia (Latitude: 37°50′ S; Longitude: 145°29′ E) was provided by Monash University and sequenced by Deakin University (Accession no.: SAMN13475009). DNA extracted from liver, heart and muscle tissues was sequenced using NovaSeq 6000 sequencer. The DNA shield is stored at Monash University with accession B50252 and the body of this animal is stored at Healesville Sanctuary with the same accession in a freezer. The raw whole genome sequence data of G. leadbeateri was retrieved from GenBank (SRR10641223). The obtained reads were analyzed based on quality using FastQC v0.11.9 (Andrews Citation2010) and the filtered reads were assembled using GetOrganelle v1.7.1 (Jin et al. Citation2020). The search tool tRNAscan-SE v2.0.6 (Lowe and Chan Citation2016) and MFannot webserver (Beck and Lang Citation2010) (https://megasun.bch.umontreal.ca/cgi-bin/dev_mfa/mfannotInterface.pl) were used to annotate the mitogenome. All the genes were manually checked using online NCBI blastn tool.
The complete circular mitochondrial genome of G. leadbeateri is 16,812 bp in length with the GC content of 39.5%. The mitogenome consists of 13 protein-coding genes (PCGs), 2 ribosomal RNA genes (12S rRNA and 16S rRNA) and 22 tRNA genes. The proportions of A, T, G, and C are 33.7, 26.8, 12.4, and 27.1%, respectively. All PCGs use start codon (ATG, ATA and ATT) and 10 PCGs have the stop codon (TAA, TAG, AGA). The PCG order (nad1, nad2, cox1, cox2, atp8, atp6, cox3, nad3, nad4L, nad4, nad5, nad6, cob) is the same as two other species, Dactylopsila trivirgata and Petaurus breviceps (Munemasa et al. Citation2006).
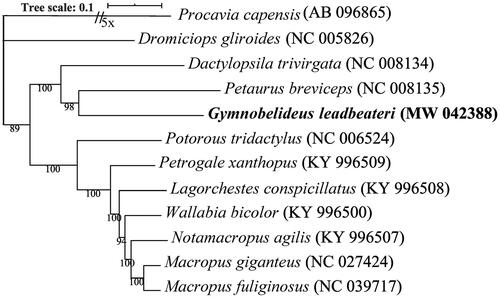
For mitogenome phylogenetic analysis, ten species were chosen from three sister families of Diprotodontia: Petauridae, Macropodidae and Potoroidae. Dromiciops gliroides and Procavia capensis were used as the outgroup taxa. The mitogenome sequences were aligned using Mafft v7.471 (Katoh and Standley Citation2013) with the –auto option, and filtered using Gblocks v0.91b (Castresana Citation2000) with block options –b4 = 5, –b5 = h. Phylogenetic tree was inferred by IQ-TREE v1.6 (Nguyen et al. Citation2015) using maximum likelihood (ML) method with the best-fit model TIM2 + F + I + G4. The resulting tree was visualized by iTOL v4 (Letunic and Bork Citation2019). Phylogenetic analyses indicate that G. leadbeateri is closely related to Petaurus breviceps and Dactylopsila trivirgata (). These data, which represent the first complete mitogenome for the G. leadbeateri provided in this study contributes to the phylogeny and protection of this Critical Endangered species.
Figure 1. Phylogenetic tree inferred from the ML analysis of the complete mitochondrial genomes of the analyzed taxa belongs to the order Diprotodontia. The bootstrap support values are given at the nodes and the species name is followed by the strain GenBank accession numbers. Isolate from present study is in bold and the scale bar represents the expected number of changes per site.
Acknowledgments
We are grateful to Dr. Alexandra Pavlova for her assistance on sample information.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data (annotated complete mitochondrial genome of Gymnobelideus leadbeateri) that support the findings of this study are submitted to NCBI GenBank (https://www.ncbi.nlm.nih.gov/) under accession number MW042388.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data; [accessed 2021 Jan 12]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Beck N, Lang B. 2010. MFannot, organelle genome annotation websever; [accessed 2021 Jan 12]. http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552.
- Harley DK. 2004. A review of recent records of Leadbeater’s possum Gymnobelideus leadbeateri. The Biology of Australian Possums and Gliders 330–338, Surrey Beatty & Sons, Chipping Norton.
- Jin J, Yu W, Yang J, Song Y, dePamphilis CW, Yi T, Li D. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47(W1):W256–W259.
- Lindenmayer DB, Cunningham RB, Tanton MT, Nix HA, Smith AP. 1991. The conservation of arboreal marsupials in the montane ash forests of the Central Highlands of Victoria, South-East Australia: III. Biol Conserv. 56(3):295–315.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Munemasa M, Nikaido M, Donnellan S, Austin CC, Okada N, Hasegawa M. 2006. Phylogenetic analysis of diprotodontian marsupials based on complete mitochondrial genomes. Genes Genet Syst. 81(3):181–191.
- Nguyen LT, Schmidt HA, Haeseler A, Minh BQ. 2015. Iq-tree: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Woinarski J, Burbidge AA. 2016. Gymnobelideus leadbeateri. The IUCN Red List of Threatened Species 2016: e.T9564A21959976.
