Abstract
The complete mitochondrial genome (mitogenome) of Mukaria splendida Distant, Citation1908 (Hemiptera: Cicadellidae: Deltocephalinae) was first reported in Mukariini. The length of this mitogenome is 16,711 bp, which has an A + T content of 79% (A = 44.5%, T = 34.5%, G = 8.8%, C = 12.2%). A total of 37 genes were annotated [13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNA), and 2 ribosomal RNA genes (rRNA)]. Among the 37 genes, 4 protein coding genes (ND1, ND4, ND4L, ND5), 8 tRNA genes (trnQ, trnC, trnY, trnF, trnH, trnP, trnL2, trnV), and 2 rRNA (12S rRNA, 16S rRNA) were encoded by N chain, and the remaining genes were encoded by J chain. Overall, there were 14 gene overlaps and 9 gene gaps in the mitochondrial genome of this species. All PCGs were started with ATD (ATA/ATT/ATG), and stopped with TAR, except ATP6, which ends with single T. The phylogenetic analysis confirms that M. splendida clustered with other Deltocephalinae species.
The genus Mukaria was established with Specimens penthimioides distance, 1908 from Sri Lankamukaria (Distant, 1908), which belongs to Cicadellidae, Deltocephalinae, and Mukariini. Up to now, only 14 species of this genus have been recorded in the world and 10 species in China, mainly distributed in the Oriental and Palearctic regions (Yang et al. Citation2016). Insects of this group are important pests of bamboo (Yao et al. Citation2019). Most of them are similar in shape, medium in size and mostly black with glossy in color. The early literature description is simple and the identification characteristic figure is incomplete or unclear, so it is difficult to identify it accurately. The mitochondrial genome was sequenced and analyzed for its structure and phylogeny of Mukaria splendida (Distant 1908) provides basic data for species identification, population genetics, and evolution of the Cicadellidae.
Total genome DNA was extracted from male adult of M. splendida using DNeasy Blood & Tissue Kit (Qiagen), samples were collected from Yuanyang County, Yunnan Province, China (102°50′44″E, 23°13′15″ N) in August 2017. Samples and genome DNA are deposited in the Institute of Entomology, Guizhou University, Guiyang, China (GUGC-IDT-00190). Sequences were sequenced by Berry Genomics on a HiSeq 2500 platform (Illumina) with 6 Gb clean data. Mitogenome was assembled using Geneious Primer (v. 2019.2.1) under reference sequence (Nephotettix cincticeps, GenBank No: NC_026977).
The complete mitogenome of M. splendida is 16,711 bp in length (GenBank No: MG813485), containing 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rRNAs), and 1 large noncoding region (Control region). In general, the M. splendida mitogenome showed obvious AT bias, which has high A + T content of 79% (A = 44.5%, T = 34.5%, G = 8.8%, C = 12.2%). The AT-skew and GC-skew are positive (0.126) and negative (−0.164). All PCGs were started with standard starting codon (ATA, ATC, ATT, ATG), and stopped with TAR (TAA/TAG), except ATP6, which stopped with single T. The length of 22 tRNAs was 1432 bp, of which the shortest was trnL1 (60 bp), and the longest was trnK (71 bp). Except for the secondary structure of trnS1 (AGN) lacks DHU arm, other 21 tRNAs was typical clover structure. Among the two rRNA genes, the 16SrRNA gene is 1205 bp in size, which was located between trnL2 and trnV, and the 12SrRNA gene is 759 bp in size, located between trnV gene and control region. There are 14 overlapping regions in M. splendida's mitochondrial genome, with a total of 95 bp. The longest overlapping region with a length of 26 bp is between trnH and ND4, and it has 9 gene spacer regions, a total of 47 bp, of which the longest interval with a length of 19 bp is between trnY and COX1.
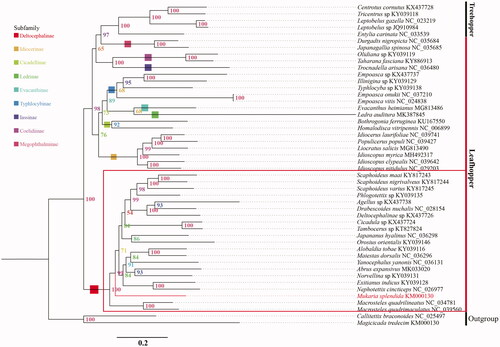
The phylogenetic relationships were analyzed based on the concatenated nucleotide sequences of 13 PCGs and 2 rRNAs from 46 Membracoidea species (41 Cicadellidae species and 5 Membracidae species) and two outgroups (Cicadidae). Each PCG and rRNA sequence was aligned using program of MACSE (Ranwez et al. Citation2011) and MAFFT (Katoh et al. Citation2005), respectively. And aligned sequences were eliminated using Gblocks 9.1 b (Talavera and Castresana Citation2007). The phylogenetic relationships of M. splendida were reconstructed with IQ-TREE using an ultrafast bootstrap approximation approach with 50,000 replicates (). The phylogenetic analysis confirms that M. splendida clustered with other Deltocephalinae species. Up to now, not a lot of molecular studies have been reported with Mukariini, and we hope that our data can be useful for further study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at: https://www.ncbi.nlm.nih.gov/nuccore/MG813485. Associated BioProject, SRA, and BioSample accession numbers are https://www.ncbi.nlm.nih.gov/bioproject/PRJNA 674714, https://www.ncbi.nlm.nih.gov/sra/SRR 12989394, and https://www.ncbi.nlm.nih.gov/biosample/SAMN 16673172, respectively.
Additional information
Funding
References
- Distant WL. 1908. Family Jassidae. The fauna of British Indian including Ceylon and Burma. Rhynchota. 4:157–419.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577.
- Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33(2):511–518.
- Ranwez V, Harispe S, Delsuc F, Douzery EJP. 2011. MACSE: Multiple Alignment of Coding SEquences accounting for frameshifts and stop codons. PLOS One. 6(9):e22594.
- Yang L, Chen XS, Li ZZ. 2016. Bambusimukaria, a new bamboo-feeding leafhopper genus from China, with description of one new species (Hemiptera, Cicadellidae, Deltocephalinae, Mukaiiini). ZooKeys. 563(563):21–32.
- Yao YL, Chen XS, Yang L. 2019. A new Bambusa chungii feeding species of the genus Mukaria Distant (Hemiptera: Cicadellidae: Deltocephalinae: Mukariini) from China. Scientia Silvae Sinicae. 55(7):105–110.