Abstract
The Egyptian mongoose, Herpestes ichneumon, is the only extant mongoose in Europe, with populations still distributed in Africa and the Middle East. In this study, we present the first mitochondrial genome sequence of Herpestes ichneumon and we investigate its phylogenetic position within Feliformia suborder. The resultant mitogenome sequence is 16,775 bps, composed of a conserved set of 37 genes containing 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a control region. Our results represent a valuable resource for further phylogeographical studies.
The Egyptian mongoose, Herpestes ichneumon (Linnaeus, 1758), is a carnivore species from the Herpestidae family widely distributed in Africa and the Middle East including Jordan, Israel, Palestinian Territories, Syria, Turkey and Lebanon (Corbet Citation1978; Tohme and Tohme Citation1985; Masseti Citation2009; Kingdon Citation2015; Özkurt Citation2015). This species is also the only mongoose living in Europe, although its presence is restricted to the Iberian Peninsula including Spain and Portugal (Delibes Citation1982; Palomares and Delibes Citation1993; Barros Citation2009; Balmori and Carbonell Citation2012).
In this study, we generated the mitochondrial genome sequence of Herpestes ichneumon using historical DNA extracted from a museum specimen and we examined its phylogenetic position within Feliformia suborder. The specimen was obtained from Akkar, a region located in the far North of Lebanon close to the Lebanese-Syrian borders. The generated sequence was submitted to GenBank database under the following accession number (MW019668). This is the first study to produce the mitochondrial genome of this species.
A footpad tissue sample was collected under sterile conditions from a museum specimen preserved in the Museum of Birds, Mammals and Butterflies of Qobayat-Lebanon (34°34′00″N, 36°16′45″E). DNA was extracted using a modified silica-column extraction protocol (McDonough et al. Citation2018) in a clean, PCR-free laboratory dedicated to ancient DNA processing at the Smithsonian Center for Conservation Genomics (CCG) in Washington, DC where the DNA is stored. The DNA extract was quantified using a Qubit® fluorometer (Life Technologies) and DNA fragment size was estimated using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA) with High Sensitivity DNA kit. We then applied the Illumina blunt-end single-tube library preparation method for degraded DNA described by Carøe et al. (Citation2018) and determined the number of index PCR cycles by performing qPCR on the library. We performed a dual indexing PCR with TruSeq-style indices (Meyer and Kircher Citation2010) using Kapa HiFi Uracil+ (Kapa Biosystems). The library was sequenced with 2 × 150 bp paired-end reads using Illumina MiSeq® platform at the CCG. In total, 1,914,038 reads were generated.
PCR duplicates and poor-quality reads were removed from the raw sequence data with prinseq-lite-0.20.4, and adapter contamination was removed using TrimGalore 0.4.1. Mitogenome assembly, consensus generation, and features annotation were performed with Geneious v9.1.2 software (Biomatters Ltd.). Quality-filtered reads were mapped to all available and verified mitogenome sequences of species from the same genus: Herpestes javanicus (AY873843; NC_006835 and KY117548) and Herpestes brachyurus (KY117547) using Geneious mapping algorithm with Medium-Low sensitivity.
The resultant mitogenome sequence length of H. ichneumon is 16,775 bp. The average sequencing depth was 43×. The sequence contains 1 control region, 2 ribosomal RNA genes, 22 transfer RNA genes and 13 protein-coding genes including ones for NADH dehydrogenase (ND1, ND2, ND3, ND4, ND4L, ND5 and ND6), ones for cytochrome c oxidase (COX1, COX2 and COX3), ATP synthase (ATP6 and ATP8) and cytochrome b gene which is typical of a vertebrate mitochondrial genome. The base composition is 33% A, 27.4% C, 13.8% G, 25.2% T and 0.6% N; the GC content is 41.2%. Due to the degraded nature of historical DNA, some gaps were observed in our assembled mitogenome sequence in the COX1 gene and the D-loop of the control region.
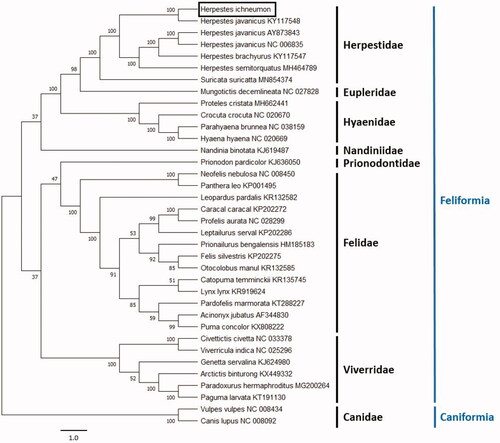
To determine the position of H. ichneumon within the Feliformia suborder and Herpestidae family, we constructed a Neighbour-Joining (NJ) tree using MEGA X (Kumar et al. Citation2018) with 1000 bootstrap replicates (Felsenstein Citation1985), including our sequence and that of several species retrieved from GenBank (). Except for Herpestes species, only one species per genus is included in the tree. Two canidae species are used as an outgroup. Evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al. Citation2004). Our phylogenetic hypothesis based on the NJ tree supports the placement of the Egyptian mongoose in the family Herpestidae. This species falls with other members of the Herpestes genus forming a sister clade to the meerkat (Suricata suricatta). Our results are in concordance with Derežanin et al. (Citation2020) study.
Figure 1. Neighbor-Joining tree of Herpestes ichneumon and other Feliformia and Caniformia species based on complete mitogenome sequences. Bootstrap support values are indicated at the base of the nodes of each clade.
Many hypotheses have been previously proposed to explain the occurrence of the Egyptian mongoose in the Iberian Peninsula, if it is an exotic species introduced in this region by Arab Muslims in the Middle Ages, or by Romans in the Roman Hispania Era, or even by Phoenicians during the 11th century BC. It has also been proposed that it colonized the Iberian Peninsula during the Late Pleistocene when sea-levels decreased (Cheylan Citation1991; Detry et al. Citation2011, Citation2018). Gaubert et al. (Citation2011) results based on cytb and control region mitochondrial fragments support the hypothesis of sweepstake dispersal of the species during Late Pleistocene sea-level fluctuations followed by long-term in situ evolution.
Our results will enable future phylogeographic studies to illuminate the reason behind the occurrence of H. ichneumon in the Iberian Peninsula and to elucidate more precisely the timing and mode of those colonization and species introduction events.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW019668.
Additional information
Funding
References
- Balmori A, Carbonell R. 2012. Expansion and distribution of the Egyptian mongoose (Herpestes ichneumon) in the Iberian Peninsula. Galemys. 24:1– 85.
- Barros T. 2009. Estatuto e distribuição (Herpestes ichneumon) do sacarrabos em Portugal [Portugal: MSc thesis]. Departamento de Biologia, Universidade de Aveiro.
- Carøe C, Gopalakrishnan S, Vinner L, Mak SS, Sinding MHS, Samaniego JA, Wales N, Sicheritz‐Pontén T, Gilbert MTP. 2018. Single-tube library preparation for degraded DNA. Methods Ecol Evol. 9(2):410–419.
- Cheylan G. 1991. Patterns of Pleistocene turnover, current distribution and speciation among Mediterranean mammals. Biogeography of Mediterranean invasions. 227–262.
- Corbet GB. 1978. The mammals of the Palaearctic region: a taxonomic review. British Museum (Natural History). Ithaca: Cornell University Press.
- Delibes M. 1982. Notas sobre la distribución pasada y actual del meloncillo Herpestes ichneumon (L.) en la Península Ibérica. Doñana Acta Vertebrata. 9:341–352.
- Derežanin L, Fickel J, Förster D. 2020. The complete mitochondrial genome of the meerkat (Suricata suricatta) and its phylogenetic relationship with other feliform species. Mitochondrial DNA B Resour. 5(1):1100–1101.
- Detry C, Bicho N, Fernandes H, Fernandes C. 2011. The Emirate of Córdoba (756–929 AD) and the introduction of the Egyptian mongoose (Herpestes ichneumon) in Iberia: the remains from Muge, Portugal. J Archaeolog Sci. 38(12):3518–3523.
- Detry C, Cardoso JL, Mora JH, Bustamante-Álvarez M, Silva AM, Pimenta J, Fernandes I, Fernandes C. 2018. Did the Romans introduce the Egyptian mongoose (Herpestes ichneumon) into the Iberian Peninsula? Naturwissenschaften. 105(11–12):63.
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39(4):783–791.
- Gaubert P, Machordom A, Morales A, López-Bao JV, Veron G, Amin M, Barros T, Basuony M, Djagoun CAMS, San EDL, et al. 2011. Comparative phylogeography of two African carnivorans presumably introduced into Europe: disentangling natural versus human-mediated dispersal across the Strait of Gibraltar. J Biogeogra. 38(2):341–358.
- Kingdon J. 2015. The Kingdon field guide to African mammals. Princeton: Bloomsbury Publishing.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Masseti M. 2009. The mongoose of the Cave of Nerja, southern Spain, is not the oldest Egyptian mongoose of Europe. Archaeofauna. 18:65–68.
- McDonough MM, Parker LD, Rotzel McInerney N, Campana MG, Maldonado JE. 2018. Performance of commonly requested destructive museum samples for mammalian genomic studies. J Mammal. 99(4):789–802.
- Meyer M, Kircher M. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010(6):pdb.prot5448–pdb.prot5448. pdb. prot5448
- Özkurt ŞÖ. 2015. Karyological and some morphological characteristics of the Egyptian mongoose, Herpestes ichneumon (Mammalia: Carnivora), along with current distribution range in Turkey. Turk J Zool. 39(3):482–487.
- Palomares F, Delibes M. 1993. Key habitats for Egyptian mongooses in Doñana National Park, South-western Spain. J Appl Ecol. 30(4):752–758.
- Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 101(30):11030–11035.
- Tohme G, Tohme H. 1985. The wild mammals of Lebanon. Lebanese University Publications. Number 16. 189p.
