Abstract
Oxalis corniculata L. is a perennial herb with a world-wide distribution. In this study, we sequenced the complete chloroplast genome of O. corniculata, which exhibited a circular genome of 155,182 bp in length with 37.5% GC content. The chloroplast genome contained a canonical quadripartite structure with a large single copy (LSC) region of 83,936 bp, a small single copy (SSC) region of 17,048 bp and a pair of 25,581 bp inverted repeats (IRs). A total of 108 unique genes, including 76 protein-coding genes (PCGs), 28 tRNA genes and four rRNA genes were found in this chloroplast genome. The phylogenetic tree was constructed based on O. corniculata and other 11 chloroplast genome sequences, which showed that O. corniculata was closely grouped with of O. corymbosa and O. drummondii.
Oxalis corniculata L., a perennial herb with unique leaf shape and bright yellow flowers, is one of the widely planted groundcover plants in the garden (Doust et al. Citation1985). In addition, the whole plant of O. corniculate is well-known as medicine, which is used in the treatment of infections, dysentery, diarrhea, skin diseases and insect bites (Raghavendra et al. Citation2006; Rehman et al. Citation2015). Here, we report the chloroplast genome of O. corniculata in the first time and aim to provide genomic resources for this species.
Total genomic DNA of O. corniculata was isolated from the fresh leaves collected in the garden of No.11 dormitory of Hainan University, China (N20°03′19.00″, E110°19′42.08″) following the standard protocol of DNA Extraction Kit (TIANGEN) and stored at −80 °C in the Key Laboratory of Genetics and Germplasm Innovation of Tropical Special Forest Trees and Ornamental Plants of Hainan University. Paired-end (150 bp) sequencing was sequenced by an Illumina HiSeq X-Ten platform. Approximately, 3.68 Gb clean data were obtained and used to assemble the complete chloroplast genomic using GetOrganelle pipeline v1.6.0 (Jin et al. Citation2018). The complete chloroplast genome of Oxalis Corymbosa (GenBank accession No. NC_048890) was used as the reference to annotate the chloroplast genome with the GeSeq (Tillich et al. Citation2017). The Geneious R11 (Kearse et al. Citation2012) was used to check the accuracy of the assembly. The complete chloroplast genome of O. corniculata had been submitted to GenBank with the accession number MT648826.
The chloroplast genome of O. corniculata has a total length of 152,146 bp with an overall GC content of 36.7%. It exhibits a canonical quadripartite structure including a large single copy (LSC) region of 83,936 bp, a small single copy (SSC) region of 17,048 bp and a pair of 25,581 bp inverted repeats (IRs). The genome encodes 108 unique genes, including 76 protein-coding genes (PCGs), 28 tRNA genes and four rRNA genes.
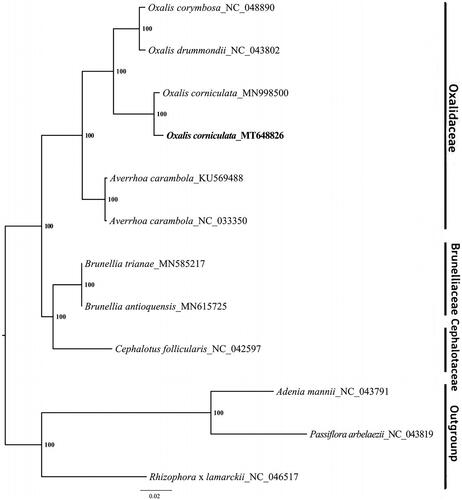
To reveal the phylogenetic position of O.corniculata, we constructed a maximum-likelihood (ML) phylogenetic tree by RAxML v.8.2.10 (Stamatakis Citation2014), which was based on 11 complete chloroplast genome sequences from the GenBank, including 8 Oxalidales species and 3 Malpighiales species as the outgroup. All the sequences were aligned by MAFFT v.7.308 (Katoh and Standley Citation2013) with default parameters. As shown in , our research illustrated that the chloroplast sequence of O. corniculata in this study was clustered closely together (96.2% identity) with the previously reported sequence of O. corniculata (Lubna et al. Citation2020). However, there were 5798 polymorphic sites between two chloroplast sequences of O. corniculata (1300 SNPs and 4498 InDels). In addition, O. corniculata was grouped with other species of the genus Oxalis such as O. corymbosa and O. drummondii. The complete chloroplast genome sequence of O. corniculata will provide an available resource for molecular identification work in this species, and contribute to the phylogenetic analysis for Oxalis genus in the future.
Figure 1. Maximum-likelihood (ML) phylogenetic tree based on complete chloroplast genome sequences of O. corniculata, five other Oxalidaceae species, two Brunelliaceae species, one Crephalotaceae species and three Malpighiales species as the outgroup. Numbers on the nodes are bootstrap values from 1000 replicates.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov/, reference number MT648826. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA659844, SRR13123621, and SAMN15929318 respectively.
References
- Doust LL, MacKinnon A, Dousp JL. 1985. Biology of Canadian Weeds: 71. Oxalis stricta L., O. corniculata L., O. dillenii Jacq. ssp. dillenii and O. dillenii Jacq. ssp. filipes (Small) Eiten. Can J Plant Sci. 65(3):691–709.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. Genome Biology. doi: 10.1186/s13059-020-02154-5.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lubna Asaf S, Jan R, Khan AL, Lee IJ. 2020. Complete chloroplast genome characterization of Oxalis corniculata and its comparison with related species from family oxalidaceae. Plants. 9(8):928. https://doi.org/10.3390/plants9080928
- Raghavendra MP, Satish S, Raveesha KA. 2006. Phytochemical analysis and antibacterial activity of Oxalis corniculata; a known medicinal plant. mySCIENCE. 1(1):72–78.
- Rehman A, Rehman A, Ahmad I. 2015. Antibacterial, antifungal, and insecticidal potentials of Oxalis corniculata and its isolated compounds. Int J Anal Chem. 2015:842468.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
