Abstract
Chesneya acaulis is a perennial herb, which restricts in Xizang (Tibet) of China, Afghanistan, and Pakistan. The complete chloroplast genome was sequenced using the Illumina Hiseq X-Ten platform. The genome lacks an inverted repeat (IR) region, containing 75 protein-coding genes, 29 tRNAs genes, and 4 rRNAs. The overall GC content is 34.6%. A phylogenetic tree based on the whole chloroplast genomes of 15 species indicated that C. acaulis had a close relationship with the genus Hedysarum, and it nested in the inverted repeat-lacking clade (IRLC), of the subfamily Papilionoideae (Leguminosae).
Chesneya acaulis (Baker) Popov, is a plateau cushion-like herb, inhabiting gravelly areas, which restricts in Xizang (Tibet) of China, Afghanistan, and Pakistan (Li Citation1981, Citation1998; Zhu and Larsen Citation2010). While in China, Chesneya acaulis only distributed in southern Xizang. Few study focused on chloroplast genome of Chesneya, a good knowledge in genomic information of Chesneya acaulis would contribute to the study of population genetics, diversity, and geological history of this genus and other plateau plants.
The fresh leaves of Chesneya acaulis were collected in Zhada County, Xizang (Tibet), China, and the voucher specimen were deposited in the herbaria of Northwest A&F University (WUK, collection #: Z.Y.Chang 2013166). Total genomic DNA were extracted from leaf tissue samples with CTAB approach (Doyle Citation1987), the genomic libraries were prepared and sequenced using the Illumina Hiseq X-Ten platform (Illumina Inc., San Diego, CA) with 150 bp paired-end reads. The resultant sequences were filtered following Yao et al. (Citation2016), the adaptor-free reads were then assembled with SPAdes 3.11 (Bankevich et al. Citation2012). The complete chloroplast (cp) genome was annotated using the Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al. Citation2004) and deposited the genome in GenBank (accession number: MW053403).
About 1.70 Gb raw reads of Chesneya acaulis were obtained, with coverage of 1441× and 128,629 bp in length. The cp genome lacked an inverted repeat (IR) region. The genome contained 75 protein-coding genes (CDS), 29 transfer RNA genes (tRNA), 4 ribosomal RNA genes (rRNA), within which 16 genes (atpF, ndhA, ndhB, trnI-AUC, trnA-UGC, petB, petD, rpl16, rpl2, trnG-UCC, rpoC1, rps12, rps12, trnL-CAA, trnV-UAC, trnK-UUU) had one intron, one gene (ycf3) had two introns. Overall GC content of the whole genome was 34.6%.
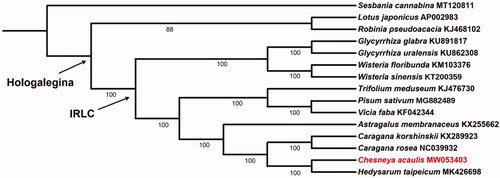
To infer the phylogenetic relationship between the newly sequenced C. acaulis and its related taxa, 14 chloroplast genomes were downloaded from GenBank and were applied to construct the systematic tree. We aligned these 15 cp genomes using MAFFT version 7 (Katoh and Standley Citation2013) and generated a maximum-likelihood (ML) tree through the program IQ-TREE version 1.4.2 (Nguyen et al. Citation2015). The result () showed that genus Chesneya had a close relationship with Hedysarum, and nested in the IR-lacking clade (IRLC), which in turn belonged to the clade of Hologalegina of papilionoid legumes as suggested by a previous study (Wojciechowski et al. Citation2004).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW053403.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li PQ. 1981. Notes on Chinese Chesneya. Acta Phytotax Sin. 19(2):235–237.
- Li PQ. 1998. Glycyrrhiza. In: Cui HB, editor. Flora reipublicae popularis sinicae. Vol. 42. Beijing: Science Press; p. 167–176.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Wojciechowski MF, Lavin M, Sanderson MJ. 2004. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am J Bot. 91(11):1846–1862.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yao X, Tan YH, Liu YY, Song Y, Yang JB, Corlett RT. 2016. Chloroplast genome structure in Ilex (Aquifoliaceae). Sci Rep. 6(1):28559.
- Zhu XY, Larsen K. 2010. Chesneya. In: Wu ZY, Hong DY, Raven PH, editors. Flora of China. Vol. 10. Beijing and St. Louis: Science Press and Missouri Botanical Garden Press; p. 500–502.