Abstract
Cirripedia is a lower crustacean that has an invaluable place in several aspects of intertidal ecology and anti-fouling research. In this study, we present the first mitochondrial genome of Chthamalus malayensis. The complete mitochondrial genome of C. malayensis is a circular molecule of 15,230 bp. In comparison to the pancrustacean ground pattern, the mitochondrial genome of C. malayensis has a deletion of the trnC gene. Phylogenetic analysis based on mitochondrial protein-coding genes showed that C. malayensis clusters with C. antennatus (BP = 98) and is grouped with C. challengeri, Octomeris sp. BKKC-2014, and Notochthamalus scabrosus. Further studies are needed to reveal the specific phylogenic relationships within Cirripedia.
Cirripedia has an invaluable place in several aspects of intertidal ecology and anti-fouling research. In this study, samples of Chthamalus malayensis were collected from the Hainan Islands (N: 20.06, E: 110.35), China. The samples were conserved in the Marine Museum of Jiangsu Ocean University. Total DNA was extracted from the muscle tissues of the samples using the TIANamp DNA Kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. DNA samples were stored at the Marine Museum of Jiangsu Ocean University (Accession number: Chma-001) and the mitochondrial genome of C. malayensis sequenced and annotated according to our previous study (Chen et al. Citation2019).
The mitochondrial genome is extranuclear genetic material that is maternal inheritance. Mitochondrial DNA can be easily sequenced and is an important molecular marker relating to metazoan phylogeny (Boore and Brown Citation1998). In this study, the mitochondrial genome of C. malayensis is presented. It is a circular DNA molecule containing 15,230 bp (GenBank accession number: MW076458), which encodes 13 protein-coding genes (PCGs), 21 tRNA genes, and 2 rRNA genes (Supplementary Table S1). In particular, a deletion of trnC exists compared with the pancrustacean ground pattern. Two PCGs (cob and nd6) and four tRNAs (trnF, trnS2, trnT, and trnQ) were found to be located in the light strand and the remaining genes were found to be located in the heavy strand. Seven longer non-coding regions with intergenic sequences more than 30 bp were found. All non-coding regions are 789 bp in length and the longest non-coding region (334 bp) is located between the 12S rRNA region and the trnK gene.
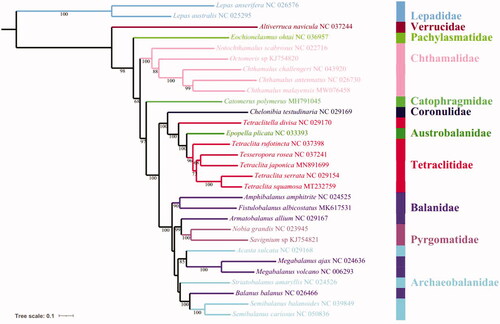
To clarify the phylogenetic relationships within Cirripedia, a phylogenetic tree was constructed based on the amino acid sequences of 13 PCGs from the complete mitochondrial genomes of 30 Cirripedia species (28 Sessilia and 2 Pedunculata) using the PhyloSuite software (Shen et al. Citation2017; Zhang et al. Citation2020). As shown in , within Chthamalidae, C. malayensis clusters with C. antennatus into a branch (BP = 98), and then grouped with C. challengeri with high support (BP = 100). Furthermore, above-mentioned three species are grouped with Octomeris sp. BKKC-2014 and Notochthamalus scabrosus, successively. The phylogenetic tree also showed that the Balanidae and Archaeobalanidae cluster together.
Figure 1. The phylogenetic tree based on 13 PCGs nucleotide acid sequences of C. malayensis and other 29 Cirripedia species.
Gene arrangements can help to better understand the phylogenetic relationships among Cirripedia species (Tsang et al. Citation2017). In comparison to the pancrustacean ground pattern, the mitochondrial genome of C. malayensis exhibits massive gene rearrangements (). In terms of Chthamalidae, C. antennatus, C. challengeri, and C. malayensis share the same gene order, however, C. malayensis has a deletion of the trnC gene. Moreover, the gene order of N. scabrosus was conserved as evidenced by its basal location in Chthamalidae. In this study, we present the first mitochondrial genome of C. malayensis. These data can help to better understand the phylogenetic history within Cirripedia, however, more data and further research are needed to reveal the phylogeny within Cirripedia.
Supplemental Material
Download MS Word (636.4 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data supporting the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW076458. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA685939, SRR13266916, and SAMN17101451, respectively.
Additional information
Funding
References
- Boore JL, Brown WM. 1998. Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr Opin Genet Dev. 8(6):668–674.
- Chen PP, Song J, Shen X, Cai YF, Chu KH, Li YQ, Tian M. 2019. Mitochondrial genome of Chthamalus challengeri (Crustacea: Sessilia): gene order comparison within Chthamalidae and phylogenetic consideration within Balanomorpha. Acta Oceanol Sin. 38(6):25–31.
- Shen X, Tsang LM, Chu KH, Chan BKK. 2017. A unique duplication of gene cluster (S2-C-Y) in Epopella plicata (Crustacea) mitochondrial genome and phylogeny within Cirripedia. Mitochondrial DNA Part A. 28(2):285–287.
- Tsang LM, Shen X, Cheang CC, Chu KH, Chan BKK. 2017. Gene rearrangement and sequence analysis of mitogenomes suggest polyphyly of Archaeobalanid and Balanid barnacles (Cirripedia: Balanomorpha). Zool Scr. 46(6):729–739.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
