Abstract
In this study, the complete chloroplast (cp) genome sequence of Ziziphus jujuba Mill. var. spinosa was mapped and determined based on Illumina sequencing data. The complete cp genome is 161,606 bp and contains a pair of inverted repeat regions of 26,479 bp each, a large single-copy region of 89,292 bp, and a small single-copy region of 19,356 bp. It harbors 112 unique genes, including 78 protein-coding genes, 4 ribosomal RNA genes, and 30 transfer RNA genes. Phylogenetic analysis based on cp genomes indicates that the cp genome of wild Z. jujuba Mill. var. spinosa is similar to that of cultivated Z. jujuba and closely related to that of Z. incurva of the family Rhamnaceae.
The Ziziphus species (family Rhamnaceae) are plants considered to have dual medicinal and food purposes, and are distributed mainly in warm and subtropical regions throughout the world (Guo et al. Citation2011). Among them, the seeds of Ziziphus jujuba Mill. var. spinosa (Bunge) Hu ex H. F. Chou have traditionally been used as an ethnomedicine in Asian countries for thousands of years (Sun et al. Citation2011; Yang et al. Citation2013). Several chloroplast (cp) DNA markers have previously been used for the diversity analysis of Z. jujuba (Huang et al. Citation2015), and the whole genome of Z. jujuba has been sequenced (Liu et al. Citation2014; Huang et al. Citation2016). Nevertheless, there exists little genomic information about Z. jujuba Mill. var. spinosa from north China. In the present study, the complete cp genome sequence of Z. jujuba Mill. var. spinosa from north China is reported based on Illumina paired-end sequencing data (GenBank accession number: MW160433).
Fresh leaves were collected from a single Z. jujuba Mill. var. spinosa tree growing in Qinglonghu town (116°5′E, 39°47′N), Fangshan District, Beijing, China. Voucher specimens (accession number: ENC850490) were deposited at the herbarium of the Beijing Academy of Forestry and Pomology Sciences. DNA extraction was performed according to a modified CTAB protocol (Li et al. Citation2013). High-throughput sequencing was carried out using a HiSeq X Ten PE150 System (Illumina, San Diego, CA, USA). The cp genome was assembled with the SPAdes 3.6.1 (Bankevich et al. Citation2012) and Sequencher 4.10 (https://www.genecodes.com/) software tools. Reference-guided assembly was then performed to reconstruct the cp genome with the BLAST program (Altschul et al. Citation1990) using closely related species as references. After filling the gaps with GapCloser (http://soap.genomics.org.cn/), a 161,606-bp cp genome was obtained for Z. jujuba Mill. var. spinosa. Annotation was performed using the Plann script (Huang and Cronk Citation2015).
The circular cp genome of Z. jujuba Mill. var. spinosa contains a pair of inverted repeat (IR) regions of 26,479 bp each, and large single-copy (LSC) and small single-copy (SSC) regions of 89,292 bp and 19,356 bp, respectively. The genome comprises 112 unique genes, including 78 protein-coding genes, 30 transfer RNA genes, and 4 ribosomal RNA genes (16S, 23S, 5S, 4.5S). Among the annotated genes, 17 genes were found to contain introns, including 15 with a single intron each and two with two introns each (clpP and ycf3).
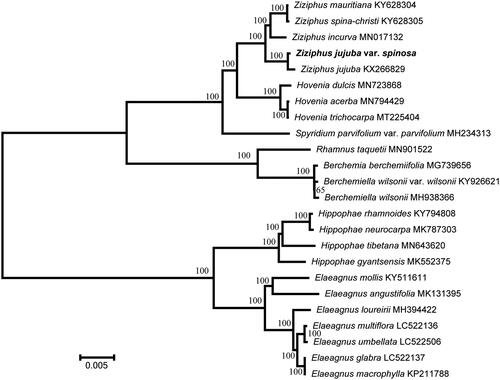
To identify the phylogenetic position of Z. jujuba Mill. var. spinosa, a maximum likelihood analysis was performed based on 24 complete chloroplast genomes using the Randomized Axelerated Maximum Likelihood (RAxML) program (Stamatakis Citation2014). The cp genome of Z. jujuba Mill. var. spinosa was found to be similar to that of cultivated Z. jujuba and closely related to that of Z. incurva of the family Rhamnaceae (). This complete cp genome can be used for subsequent population and cp genetic engineering studies, and especially to determine the phylogenetic position of Z. jujuba Mill. var. spinosa in Ziziphus Mill.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW160433.The associated BioProject, SRA, and Bio-Sample numbers are PRJNA684953, SRR13254568, SAMN17073062, respectively.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Guo S, Duan J-a, Tang Y, Qian Y, Zhao J, Qian D, Su S, Shang E. 2011. Simultaneous qualitative and quantitative analysis of triterpenic acids, saponins and flavonoids in the leaves of two Ziziphus species by HPLC-PDA-MS/ELSD. J Pharm Biomed Anal. 56(2):264–270.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Huang J, Yang X, Zhang C, Yin X, Liu S, Li X. 2015. Development of chloroplast microsatellite markers and analysis of chloroplast diversity in Chinese Jujube (Ziziphus jujuba Mill.) and Wild Jujube (Ziziphus acidojujuba Mill.). PLoS One. 10(9):e0134519.
- Huang J, Zhang C, Zhao X, Fei Z, Wan K, Zhang Z, Pang X, Yin X, Bai Y, Sun X, et al. 2016. The Jujube genome provides insights into genome evolution and the domestication of sweetness/acidity taste in fruit trees. PLoS Genet. 12(12):e1006433.
- Li J, Wang S, Yu J, Wang L, Zhou S. 2013. A modified CTAB protocol for plant dna extraction. Chin Bull Bot. 48:72–78.
- Liu MJ, Zhao J, Cai QL, Liu GC, Wang JR, Zhao ZH, Liu P, Dai L, Yan G, Wang WJ, et al. 2014. The complex jujube genome provides insights into fruit tree biology. Nat Commun. 5:5315.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Sun Y-F, Liang Z-S, Shan C-J, Viernstein H, Unger F. 2011. Comprehensive evaluation of natural antioxidants and antioxidant potentials in Ziziphus jujuba Mill. var. spinosa (Bunge) Hu ex H. F. Chou fruits based on geographical origin by TOPSIS method. Food Chem. 124(4):1612–1619.
- Yang B, Yang H, Chen F, Hua Y, Jiang Y. 2013. Phytochemical analyses of Ziziphus jujuba Mill. var. spinosa seed by ultrahigh performance liquid chromatography-tandem mass spectrometry and gas chromatography-mass spectrometry. Analyst. 138(22):6881–6888.