Abstract
The complete chloroplast (cp) genome of Actinidia latifolia was sequenced and assembled using Illumina pair-end sequencing data. The cp genome is 157,021 bp in length and comprises a large single copy (LSC) region of 88,557 bp and a small single copy (SSC) region of 21,562 bp separated by two inverted repeat (IR) regions of 23,451 bp. A total of 113 unique genes were identified, including 79 protein-coding genes, 30 tRNA genes, and four rRNA genes. Phylogenetic analysis based on cp genomes of 20 Actinidiaceae species revealed that A. latifolia was evolutionarily close to A. eriantha, A. styracifolia, and A. fulvicoma.
Kiwifruit (Actinidia Lindl.), a fruit genus with important economic and nutritional value, contains more than 50 species. As the frequent interspecific hybridization, species within Actinidia are highly variable in morphological characteristics, and their interspecific relationships are still confusing (Huang et al. Citation2013). Actinidia latifolia (Gardner & Champ.) Merr. var. latifolia is a representative Actinidia species that is widely distributed in relatively low latitude regions. It is characterized by the most flowers and fruits in an inflorescence of Actinidia, and the remarkably high vitamin C content in fresh fruit (Huang et al. Citation2013). Knowledge of the complete cp genome and phylogenetic analysis will contribute greatly to reveal the phylogeny status of A. latifolia and provide new insights into the evolutionary relationship and the taxonomy of Actinidia.
Plant material of A. latifolia was collected from Xunwu county (24°57′35″N, 115°34′51″E) of Jiangxi province, China. A specimen was deposited in the Herbarium of Lushan Botanical Garden, Chinese Academy of Sciences, under the voucher number LBG00148295. Total genomic DNA was extracted from fresh leaves using a modified CTAB method (Doyle and Doyle Citation1987) and then sequenced using the Illumina Hiseq 2000 platform. In total, 7.45 G raw reads were obtained, quality-trimmed and assembled against the plastome of A. rufa (GenBank: NC_039973.1) using the program NOVOPlasty (Dierckxsens et al. Citation2016). The annotated cp genome was submitted to GenBank with assigned accession number of MW057417.
The complete chloroplast genome of A. latifolia is 157,021 bp in length. It consists of two inverted repeat (IR) regions of 23,451 bp separated by the large single-copy (LSC) region of 88,557 bp and small single-copy (SSC) region of 21,562 bp. The overall GC content of the genome is 37.18%, and the GC content of the LSC, SSC, and IRa/b regions is 35.45%, 31.29%, and 43.14%, respectively. The cp genome possesses 113 unique functional genes, including 79 protein-coding genes, 30 distinct tRNA genes, and four rRNA genes. Five protein-coding genes, eight tRNA genes, and all four rRNA genes were totally duplicated in the IR regions. Of the 113 unique genes, six tRNA genes and seven protein-coding genes contain a single intron, and one gene (ycf3) has two introns.
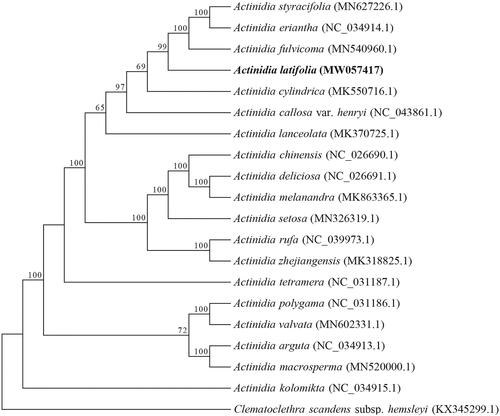
We reconstructed a phylogenetic tree based on 20 complete cp genome sequences of Actinidiaceae. The maximum likelihood (ML) tree was constructed using RAxML8.0 (Stamatakis Citation2014) with 100 bootstrap replicates under the GTR + G model. The tree topography was generally congruent with previous reports based on genome-wide SNPs (Liu et al. Citation2017) and chloroplast matK gene (Li et al. Citation2002). The evolutionary analysis showed that all species with smooth-skinned fruit, such as A. kolomikta, A. valvata, and A. macrosperma, were located at the relatively basal positions of Actinidia, and A. latifolia was evolutionarily close to A. eriantha, A. styracifolia, and A. fulvicoma (). The complete plastome can be subsequently used for genetic diversity studies of A. latifolia and will be benefit for future studies on phylogeny and taxonomy of Actinidia.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW057417.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Huang HW, Zhong CH, Jiang ZW, Li XW, Yao XH, Li DW, Wang SM, Li ZZ, Gong JJ, Liu YF, et al. 2013. Actinidia: taxonomy, resources, domestication, cultivation. Beijing: Science Press.
- Li J, Huang H, Sang T. 2002. Molecular phylogeny and infrageneric classification of Actinidia (Actinidiaceae). Syst Bot. 27(2):408–415.
- Liu Y, Li D, Zhang Q, Song C, Zhong C, Zhang X, Wang Y, Yao X, Wang Z, Zeng S, et al. 2017. Rapid radiations of both kiwifruit hybrid lineages and their parents shed light on a two-layer mode of species diversification. New Phytol. 215(2):877–890.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.