Abstract
Achyranthes bidentata is a popular Chinese medicine, that has its place in the treatment of spasm, osteodynia of the lumbar region and knees. The complete chloroplast (cp) genome of A. bidentata was determined. The complete cp genome is 151,543 bp in length and includes a large single-copy (LSC) region of 83,922 bp, a small single-copy (SSC) region of 17,251 bp, and a pair of inverted repeat regions (IRs) of 25,185 bp. It encodes 131 genes, including 86 protein-coding genes, eight rRNAs, and 37 tRNAs. Phylogenetic analysis indicates that four samples of A. bidentata formed a clade with a 100% bootstrap value.
Achyranthes bidentata Blume (Amaranthaceae), a perennial herbaceous plant, is widely distributed and grown in China (Wang et al. Citation2013). As a popular Chinese medicine, A. bidentata has its place in the treatment of spasm, osteodynia of the lumbar region and knees, and flaccidity of limbs owning to its medical functions of dissipating blood stasis, nourishing the liver and kidney, and strengthening the bones and muscles (Shen et al. Citation2011). It is therefore necessary to study this plant in detail. The advent of high-throughput sequencing technology makes it possible to acquire large amounts of genome data rapidly (He et al. Citation2017). In this study, we report the complete chloroplast (cp) genome of A. bidentata. The raw sequence data have been deposited into NCBI SRA with project accession of PRJNA662739, and the plastome sequence has been was registered in GenBank with the accession number MT955652.
The plant material of A. bidentata was sampled from the Renhe Community garden in Kaifeng city, China (34°49′8.4″N, 114°23′2.4″E), its voucher specimen (HENU20200523) has been kept at the Herbarium of Henan University. Total genomic DNA was extracted from leaf tissue with the SDS method and high-quality DNA was randomly fragmented to 350 bp. The paired-end library was then sequenced using Illumina NovaSeq PE150 at Beijing Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). Approximately, 5-Gb raw data were generated, and these were trimmed and assembled into contigs using the CLC Genomics Workbench 9.5.2 (CLC Inc., Aarhus, Denmark). The complete cp genome of A. bidentata was later constructed using Amaranthus caudatus (Amaranthaceae, GenBank accession number: NC_040143) as a reference sequence and annotated using Geneious R11 (Biomatters, Auckland, New Zealand), as described by Liu et al. (Citation2017, Citation2018).
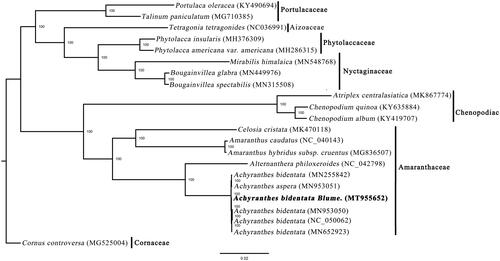
Figure 1. Phylogenetic relationships of Lamiales inferred based on whole chloroplast genome sequences. Number above each node indicates the ML bootstrap support values.
The complete plastome of A. bidentata is 151,543 bp in length and includes a large single-copy (LSC) region of 83,922 bp, a small single-copy (SSC) region of 17,251 bp, and a pair of inverted repeat regions (IRa and IRb) of 25,185 bp. The whole cp genome encoded 131 genes, including 86 protein-coding genes (PCGs), 37 tRNAs, and four rRNA operons. Among these genes, 14 genes (atpF, ndhA, ndhB, petB, petD, rpoc1, rpl16, rps16, trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) harbored one intron and two genes (clpP and ycf3) had two introns. Most genes occurred in a single copy; however, six PCG genes (ndhB, rpl2, rpl23, rps7, ycf2, and ycf15), seven tRNA genes (trnA-UGC, trnH-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and four rRNA genes (rrn4.5, rrn5, rrn16, and rrn23) in the IR regions were duplicated. The overall AT content of the cp genome is 61.9% and the corresponding values in LSC, SSC, and IR regions are 65.8, 70.0, and 57.5%, respectively. The nucleotide similarity of A. bidentata among the sequence we constructed and the others (MN652923, MN255842, MN953050, NC_050062) reported on NCBI is 99.8%, and there are 160 SNPS and 19 InDel in these sequences.
The phylogenetic tree, including A. bidentata and 16 other species in Caryophyllales, was constructed based on whole plastome sequences using RAxML-HPC XSEDE v.8.2.8 from CIPRES (http://www.phylo.org/) (Stamatakis Citation2014), with Cornus controversa as the outgroup. The tree showed four samples of A. bidentata formed a clade with a 100% bootstrap value (). This result will provide valuable insight into conservation and evolutionary histories for this important species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov. The reference numbers of the raw sequence data and the genome are PRJNA662739 and MT955652, respectively.
Additional information
Funding
References
- He Y, Yuan W, Dong M, Han Y, Shang F. 2017. The first genetic map in sweet osmanthus (Osmanthus fragrans lour.) using specific locus amplified fragment sequencing. Front Plant Sci. 8:1621.
- Liu LX, Li R, Worth JRP, Li X, Li P, Cameron KM, Fu CX. 2017. The complete chloroplast genome of Chinese bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of Fagales. Front Plant Sci. 8:968.
- Liu LX, Li P, Zhang HW, Worth JRP. 2018. Whole chloroplast genome sequences of the Japanese hemlocks, Tsuga diversifolia and T. sieboldii, and development of chloroplast microsatellite markers applicable to East Asian Tsuga. J Forest Res. 23(5):318–323.
- Shen Y, Zhang Q, Gao X, Ding F. 2011. An active fraction of Achyranthes bidentata polypeptides prevents apoptosis induced by serum deprivation in SH-SY5Y cells through activation of PI3K/Akt/Gsk3β pathways. Neurochem Res. 36(11):2186–2194.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang Y, Shen W, Yang L, Zhao H, Gu Y, Yuan Y. 2013. The protective effects of Achyranthes bidentata polypeptides on rat sciatic nerve crush injury causes modulation of neurotrophic factors. Neurochem Res. 38(3):538–546.
