Abstract
Puccinellia distans is a perennial gramineous plant with the characteristics of drought and salt tolerance. It is a special pioneer plant for saline-alkali land improvement and is increasingly used for ecological restoration of saline-alkali grassland. However, the evolutionary relationship of P. distans is limited in study. In this study, the complete chloroplast genome sequence of P. distans was evaluated. The complete chloroplast genome of P. distans was 135,647 bp in length, containing a pair of inverted repeated (IR) regions (21,444 bp) that are separated by a large single-copy (LSC) region of 800,15 bp, and a small single-copy (SSC) region of 12,744 bp. A total of 129 functional genes were annotated, including 83 protein-coding genes (mRNA), 38 tRNA genes, and 8 rRNA genes. The phylogenetic relationships of 12 species indicated that P. distans was closely related to P. muttalliana. This complete chloroplast genome will provide a theoretical basis for species identification and biological research.
Puccinellia distans is a perennial, monocotyledonous C3 plant of Poaceae family, mainly distributed in the Northeast and Northwest of China (Elisabetta et al. Citation2019). It is an excellent forage grass with the characteristics of early turning green, high nutritional value, and good palatability (Sun Citation2014). In addition, P. distans is a special pioneer plant for saline-alkali soil improvement because of its excellent drought and salt tolerance (Zhang et al. Citation2013). It is gradually used for ecological restoration (Peng et al. Citation2004). However, the evolutionary relationship of P. distans is limited. In this study, the complete chloroplast genome of P. distans was sequenced on the Illumina NovaSeq platform (Genbank accession number: MW044608), which would provide a theoretical basis for species identification and biological research in future study.
The fresh leave samples of P. distans were collected in experiment station of perennial forage in Xihai Town, Haibei Prefecture, Qinghai, China (36°49′297″N, 101°45′266″E). The voucher specimen was kept in Herbarium of Low-temperature library of Germplasm resources in Qinghai Academy of Animal and Veterinary Sciences (09-129, contact person and email are Xiaoxing Wei, [email protected]). The total genomic DNA of P. distans was extracted from the fresh leaves with a modified CTAB method (Li et al. Citation2018). One library was constructed using PCR amplification. The template size is 300 bp. The genome sequencing was performed with an Illumina NovaSeq platform (Genepioneer Biotechnologies, Nanjing, China). The trimmed reads were mainly assembled using SPAdes (Bankevich et al. Citation2012). The assembled genome was annotated using Cp GAVAS (Liu et al. Citation2012).
The complete chloroplast genome of P. distans was 135,647 bp in length, containing a pair of inverted repeated (IR) regions (21,444 bp) that are separated by a large single copy (LSC) region of 800,15 bp, and a small single copy (SSC) region of 12,744 bp. The GC content of the whole chloroplast genome was 38.32%. A total of 129 functional genes were annotated, including 83 protein-coding genes (mRNA), 38 tRNA genes, and 8 rRNA genes. The protein-coding genes, tRNA genes, and rRNA genes account for 64.34%, 29.46%, and 6.20% of all annotated genes, respectively.
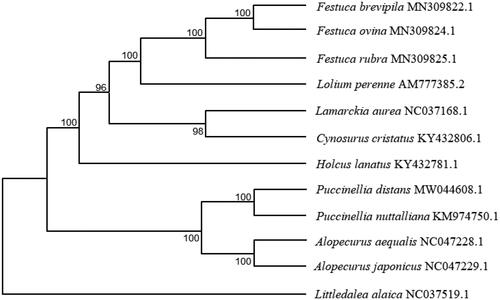
To reveal the phylogenetic position of P. distans, a maximum likelihood (ML) phylogenetic tree () was constructed with MEGA version 7 (Huang et al. Citation2020) using the coding sequences of 12 species. All the 12 species are Gramineae and belong to different genera. The results showed that P. distans was closely related to P. muttalliana. This study will contribute to its future breeding and biological research.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no.MW044608. The associated Bio-Project, SRA, and Bio-Sample numbers are PRJNA680621, SRR13375866, and SAMN16898198 respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Elisabetta O, Gianni R, Francesca G. 2019. Effects of foliar application of glycine betaine and chitosan on Puccinellia distans (Jacq.) Pari, subjected to salt stress. Biologia Futura. 70(1):47–55.
- Huang YL, Sun XQ, Jiang J, Fei YL, Hou SL. 2020. The complete mitochondrial genome sequence of Centropus bengalensis (Lesser Coucal). Mitochondrial DNA Part B. 5(2):1236–1237.
- Li X, Li YF, Zang MY, Li MZ, Fang YM. 2018. Complete chloroplast genome sequence and phylogenetic analysis of Quercusacutissima. IJMS. 19(8):2443–2459.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Lin XH, Guan XJ. 2012. Cp GAVAS, an integrated web server for the annotation, visualization, analysis, and Gen Bank submission of completely sequenced chloroplast genome sequences. BMC Genoms. 13(1):715.
- Peng YH, Zhang YF, Mao YQ, Wang SM, Su WA, Tang C. 2004. Alkali grass resists salt stress through high [K+] and an endodermis barrier to Na++. J Exp Bot. 55(398):939–949.
- Sun B. 2014. Exogenous additives on the seed germination and seedling growth of Puccinellia distans and Nitraria tangutorum Bobrunder salt-stress [master thesis]. Taian, China: Shandong Agriculture University.
- Zhang HN, Zhou QP, Yan HB, Liang GL, Liu WH. 2013. Effects of salt stress on germination of five Puccinellia distans materials. Pratacult Sci. 30(11):1767–1770.