Abstract
The recently published mitogenome of milk shark Rhizoprionodon acutus (MN602076/NC_046016) was fully resolved in an unexpected phylogenetic position in the original mitogenome announcement, which rendered the genus Scoliodon paraphyletic. Here, we show that this mitogenome is actually that of a misidentified Pacific spadenose shark (Scoliodon macrorhynchos). The error is documented to avoid the perpetuation of erroneous sequence information in the literature.
Introduction
The milk shark Rhizoprionodon acutus (Rüppell, 1837) is widely distributed in coastal areas ranging from the tropical east Atlantic Ocean, to the Indian Ocean and western Pacific Ocean (Compagno Citation1984). The first complete mitochondrial genome (hereafter mitogenome) of this species and genus was published by Liu et al. (Citation2019) based on an individual collected in the Beibu Gulf, China (GenBank accession number MN602076, RefSeq number NC_046016). Their mitogenome announcement included a maximum-likelihood cladogram based on 13 protein coding genes which placed the R. acutus sequence in a fully supported clade sister to the Pacific spadenose shark (Scoliodon macrorhynchos) and together these were sister to Indian spadenose shark (S. laticaudus). The latter result indicates paraphyly of Scoliodon. These results are surprising because the two recognized species of Scoliodon are so similar morphologically that they were treated as a single species until recently (White et al. Citation2010). In contrast, Rhizoprionodon acutus is rather distinct and differs from the two species of Scoliodon by the shape of its snout, caudal peduncle and pectoral fins, and in the position of its first dorsal fin (Springer Citation1964; White et al. Citation2010). Using a strategy described by Botero-Castro et al. (Citation2016) and expanded by Sangster and Luksenburg (Citation2020), we show that the accession MN602076 is a mitogenome sequence of S. macrorhynchos.
Materials and methods
We verified the identity of MN602076 by performing a phylogenetic analysis using sequences from its three protein-coding genes (PCGs): NADH dehydrogenase subunit 2 (ND2, 1047 bp), cytochrome oxidase subunit I (COI, 655 bp), and NADH dehydrogenase subunit 4 (ND4) plus adjacent tRNA-His, tRNA-Ser, and partial tRNA-Leu genes (873 bp). These are among the most commonly used mitochondrial markers in systematic studies of Chondrichthyes, and were selected because sufficient numbers of reference sequences were available.
A phylogeny was also constructed using sequences of the 13 PCGs included in the mitogenome but with the data set trimmed by GBLOCKS (Castresana Citation2000). GBLOCKS eliminates poorly aligned positions and divergent regions, which may not be homologous or may have been saturated by multiple substitutions (Castresana Citation2000). This resulted in an alignment of 11,374 bp. The MITOS2 web server (Bernt et al. Citation2013) was used to obtain information on the first and last positions of individual genes. CLUSTALW (as implemented in MEGA7, Kumar et al. Citation2016) was used to align sequences. Maximum-likelihood phylogenies were obtained using MEGA7. The appropriate substitution model for each data set was selected using the Akaike information criterion. In all cases, the GTR + G+I model was selected. Sequence divergence was calculated as uncorrected p values with complete deletion of nucleotide positions with missing data.
Results
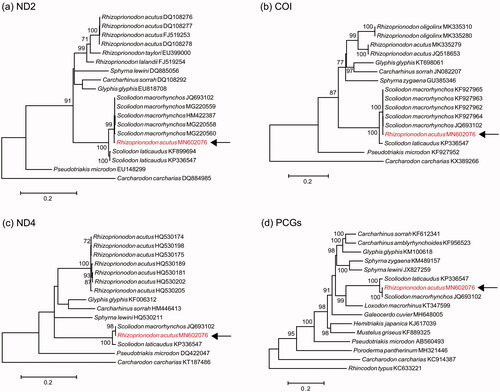
In all analyses, MN602076 clustered with S. macrorhynchos with high bootstrap support (98–100%), rather than with other sequences of R. acutus (). In addition, MN602076 was extremely similar to JQ693102 (Chen et al. 2014). Sequence divergence of the entire mitogenome of MN602076 and that of S. macrorhynchos (JQ693102) was 0.2%, whereas that of S. macrorhynchos and S. laticaudus was 3.6%. No bona fide mitogenome of R. acutus is available for comparison, but sequence divergence in ND2 and COI between MN602076 and R. acutus was substantial (ND2: 15.8–15.9%; COI: 11.7–11.9%).
Figure 1. ML phylogenies of carcharhiniform sharks and outgroup(s) based on (a) COI (655 bp), (b) ND2 (1047 bp), (c) ND4 (873 bp), and (d) mitogenomes (13 PCGs, trimmed with GBLOCKS; 11,374 bp). Numbers along branches represent bootstrap support values (>70%) based on 1000 pseudoreplications. Note the consistent placement of MN602076 with Scoliodon macrorhynchos.
In a gene tree of >12,000 COI sequences of Chondrichthyes (data not shown), several sequences identified as ‘Etmopterus lucifer’ (EF607379, EF607380, EF607381, EU595124) and ‘Etmopterus sp.’ (EU595125, EU595126, EU595127, EF607378) also clustered with S. macrorhynchos and MN602076. The eight sequences were all deposited on GenBank by the same researcher (J. Zhang), and four of these were published in Zhang (Citation2011). These sequences did not cluster with 23 other sequences of E. lucifer and were clearly misidentified. Thus, these were excluded from the formal analysis (). A COI sequence identified as ‘S. macrorhynchos’ (HM422387) clustered among S. laticaudus (data not shown) and was considered to have been either misidentified or perhaps a hybrid. This sequence was also excluded from the formal analysis.
Discussion
Our results show that MN602076, originally attributed to R. acutus, differs from multiple mitochondrial DNA sequences of R. acutus and is nearly identical to that of S. macrorhynchos. The most likely explanation for this is that the original specimen was misidentified and represents an individual of S. macrorhynchos.
An alternative explanation for the observed similarity of MN602076 to S. macrorhynchos is hybridization. Hybridization may result in the transfer of a mitochondrial genome of one species into organisms phenotypically similar to another species. Hybridization is known to have occurred in blacktip sharks (genus Carcharhinus; Morgan et al. Citation2012), dusky and Galapagos sharks (genus Carcharhinus; Corrigan et al. Citation2017; Pazmiño et al. Citation2019), and hammerhead sharks (genus Sphyrna; Barker et al. Citation2019). However, these cases represent closely related members of the same genus. Hybridization of R. acutus and S. macrorhynchos has never been recorded and would seem unlikely given their large genetic divergence (>11%) and non-sister relationship (Vélez-Zuazo and Agnarsson Citation2011; Naylor et al. Citation2012; Chen and Kishino Citation2015). Detailed morphological comparisons, or analysis of nuclear DNA data of the relevant specimen and the two potential parental species, is necessary to demonstrate hybridization. In the absence of such data, and in view of the lack of documented cases on intergeneric hybridization in sharks, we regard MN602076 as the mitogenome of a misidentified S. macrorhynchos.
The mitogenome announcement that described MN602076 (Liu et al. Citation2019) presented its phylogenetic position in a cladogram (Liu et al. Citation2019). This practice hides the true branch lengths (i.e. the number of substitutions along each branch), and prevents the detection of branches that are suspiciously long (e.g. resulting from sequencing errors or chimerism) or suspiciously short (e.g. resulting from misidentification). Thus, we reiterate the advice of Botero-Castro et al. (Citation2016) to present phylogenetic relationships as phylograms.
Reporting misidentifications is necessary because the accumulation of erroneous sequences compromises downstream applications, including – but not limited to – DNA identification, primer design for intraspecific studies, phylogenetic inference, historical biogeography, taxonomy, and comparative analysis. In addition to MN602076, we found misidentified COI sequences of S. laticaudus (one listed as ‘S. macrorhynchos’) and S. macrorhynchos (eight listed as ‘Etmopterus lucifer’ or ‘Etmopterus sp.’). We urge other scientists to help keep the field of mitogenomics healthy and trustworthy by exposing and reporting misidentified or otherwise problematic sequences.
Acknowledgements
We are grateful to Jeffery Hughey and three anonymous reviewers for offering suggestions that improved the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study were published previously and are openly available on GenBank at https://www.ncbi.nlm.nih.gov/nucleotide.
References
- Barker AM, Adams DH, Driggers WB, Frazier BS, Portnoy DS. 2019. Hybridization between sympatric hammerhead sharks in the Western North Atlantic Ocean. Biol Lett. 15(4):20190004.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Botero-Castro F, Delsuc F, Douzery EJ. 2016. Thrice better than once: quality control guidelines to validate new mitogenomes. Mitochondrial DNA Part A. 27(1):449–454.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552.
- Chen H, Kishino H. 2015. Global pattern of phylogenetic species composition of shark and its conservation priority. Ecol Evol. 5(19):4455–4465.
- Chen X, Peng X, Zhang P, Yang S, Liu M. 2014. Complete mitochondrial genome of the spadenose shark (Scoliodon macrorhynchos). Mitochondrial DNA. 25(2):91–92.
- Compagno LJV. 1984. FAO species catalogue. Vol. 4. Sharks of the world. An annotated and illustrated catalogue of shark species known to date. Part 2 – Carcharhiniformes. FAO Fisheries Synopsis No. 125(4/2). Rome: FAO; p. 251–655.
- Corrigan S, Delser PM, Eddy C, Duffy C, Yang L, Chenhong L, Bazinet AL, Mona S, Naylor GJP. 2017. Historical introgression drives pervasive mitochondrial admixture between two species of pelagic sharks. Mol Phylogenet Evol. 110(1):122–126.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Liu Y, Shan B, Yang C, Zhao Y, Liu M, Xie Q, Sun D. 2019. The complete mitochondrial genome of milk shark, Rhizoprionodon acutus (Ruppell 1837). Mitochondrial DNA Part B. 5(1):310–311.
- Morgan J, Harry A, Welch D, Street R, White J, Geraghty P, Macbeth W, Tobin A, Simpfendorfer C, Ovenden J. 2012. Detection of interspecies hybridisation in Chondrichthyes: hybrids and hybrid offspring between Australian (Carcharhinus tilstoni) and common (C. limbatus) blacktip shark found in an Australian fishery. Conserv Genet. 13(2):455–463.
- Naylor GJP, Caira JN, Jensen K, Rosana KA, Straube N, Lakner C. 2012. Elasmobranch phylogeny: a mitochondrial estimate based on 595 species. In: Carrier JC, Musick JA, Heithaus MR, editors. The biology of sharks and their relatives. Boca Raton, FL, USA: CRC Press; p. 31–56.
- Pazmiño DA, van Herderden L, Simpfendorfer CA, Junge C, Donnellan SC, Hoyos-Padilla EM, Duffy CAJ, Huveneers C, Gillanders BM, Butcher PA, et al. 2019. Introgressive hybridisation between two widespread sharks in the East Pacific region. Mol Phylogenet Evol. 136(1):119–127.
- Sangster G, Luksenburg JA. 2020. The published complete mitochondrial genome of Eptesicus serotinus is a chimera of Vespertilio sinensis and Hypsugo alaschanicus (Mammalia: Chiroptera). Mitochondrial DNA Part B. 5(3):2661–2664.
- Springer VG. 1964. A revision of the carcharhinid shark genera Scoliodon, Loxodon, and Rhizoprionodon. Proc US Nat Mus. 115(3493):559–632.
- Vélez-Zuazo X, Agnarsson I. 2011. Shark tales: a molecular species-level phylogeny of sharks (Selachimorpha, Chondrichthyes). Mol Phylogenet Evol. 58(2):207–217.
- White WT, Last PR, Naylor GJP. 2010. Scoliodon macrorhynchos (Bleeker, 1852), a second species of spadenose shark from the Western Pacific (Carcharhiniformes: Carcharhinidae). In: Last PR, White WT, Pogonoski JJ, editors. Descriptions of new sharks and rays from Borneo. CSIRO Marine and Atmospheric Research Paper No. 32, p. 61–76. http://hdl.handle.net/102.100.100/107848?index=1
- Zhang J. 2011. Species identification of marine fishes in china with DNA barcoding. Evid Based Complement Alternat Med. 2011:978253.
