Abstract
The complete mitogenome of Nasimyia megacephala Yang & Yang, Citation2010 (Stratiomyidae: Pachygastrinae) successfully sequenced. The mitogenome of N. megacephala is a circular DNA molecule of 16,069 bp in length with 72% AT content, consisting of 13 PCGs, two rRNAs, 22 tRNAs, and a large control region. The gene order is consistent with other dipteran mitogenomes. The phylogenetic tree was reconstructed using maximum likelihood analysis, and the topology revealed that Pachygastrinae is not a monophyletic group.
The genus Nasimyia was erected by Yang and Yang in 2010 (Yang and Yang Citation2010). It is a small genus of the subfamily Pachygastrinae of Stratiomyidae with four species described (Yang et al. Citation2013), and all four species were recorded in China. Nasimyia megacephala is the type species of Nasimyia. Here, we analyzed the mitogenome of N. megacephala to provide insights into the phylogenetic relationship of Stratiomyidae.
Adult individuals of N. megacephala (GZAF-2020-DS1442) were obtained in Luodian County (E106.7917, N25.4075), Guizhou province, China in May 2020, and were stored in Insect Herbarium of Guizhou Academy of Forestry (URL, Zaihua Yang and [email protected]), Guiyang. Total genomic DNA was extracted from the specimen’s thoracic muscle using the DNeasy Blood & Tissue Kits (Qiagen, Valencia, CA). Genomic DNA was sequenced by the high-throughput Illumina Hiseq 4000 platform. Next, raw data assembled using NOVOPlasty v2.7.0 (Dierckxsens et al. Citation2017) with cox1 sequence from Nemotelus notatus (GenBank accession no. MT584142) as the initial seed. Finally, the assembled data was annotated by MITOZ v1.04 (Meng et al. Citation2019).
The mitogenome of N. megacephala with a length of 16,069 bp was submitted to GenBank (Genbank accession no. MW115118). All of the 37 typical animal mitochondrial genes (13 PCGs, 22 tRNAs, and two rRNAs) and a control region were identified. All genes display the same order as the putative ancestral arrangement of insects (Clary and Wolstenholme Citation1985). The AT content of the mitogenome is 72% (37.5% A, 17.7% C, 10.2% G, 34.5% T), indicating significant AT bias. Thirteen PCGs were found to be 11,193 bp in size, accounting for 69.7% of the entire mitogenome. In 13 PCGs, cox1 starts with TCG, while atp6, atp8, cox2, cox3, cytb, nad1, nad2, nad3, nad4L, nad4, nad5, and nad6 share typical start codon ATT/G. Furthermore, all PCGs use TAA/G as the termination codon. The newly-sequenced mitogenome has the complete 22 tRNA genes, ranging from 64 to 72 bp in length. The non-coding control region was detected between rrnS and trnI is 1,205 bp long with an AT content of 86.1%.
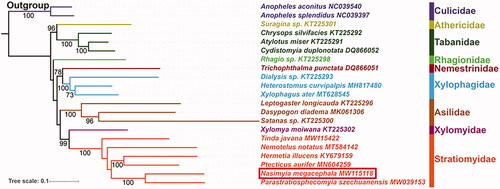
For nucleotide sequence data of 13 PCGs, the best partitioning scheme and nucleotide substitution model for maximum likelihood (ML) phylogenetic analyses were determined with PartitionFinder2 (TIM + I + G for Subset1 (atp6 and nad3), GTR + I + G for Subset2 (atp8, nad2, and nad6), Subset3 (cox1, cox2, cox3, and cytb), and Subset4 (nad1, nad4L, nad4, and nad5)) (Lanfear et al. Citation2017). The phylogenetic tree was reconstructed using IQ‒TREE v1.6.3 (Nguyen et al. Citation2015) with ultrafast bootstrap (UFB) approximation approach () (Minh et al. Citation2013). Bootstrap support (BS) values were calculated with 10,000 replicates. The phylogenetic relationships among Brachycera is recovered as ((Tabanidae + Athericidae) + (Rhagionidae + ((Xylophagidae + Nemestrinidae) + (Asilidae + (Stratiomyidae + Xylomyidae))))), which is similar to previously published phylogenies (Zhou et al. Citation2017; Ding and Yang Citation2020). But our analyses include more taxon sample. Besides, the phylogenetic tree also shows that each family is the monophyletic group with high support (BS = 100). In Stratiomyidae, the monophyly of Pachygastrinae (including Nasimyia megacephala, Parastratiosphecomyia szechuanensis, and Tinda javana in this study) is not supported.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW115118. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA693466, SRR13498970, and SAMN17388037, respectively.
Additional information
Funding
References
- Clary DO, Wolstenholme DR. 1985. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 22(3):252–271.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Ding SM, Yang D. 2020. Complete mitochondrial genome of Coenomyia ferruginea (Scopoli). (Diptera, Coenomyiidae). Mitochondrial DNA B Resour. 5(3):2711–2712.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Yang ZH, Hauser M, Yang MF, Zhang TT. 2013. The Oriental genus Nasimyia (Diptera: Stratiomyidae): geographical distribution, key to species and descriptions of three new species. Zootaxa. 3619(5):526–540.
- Yang ZH, Yang MF. 2010. A new genus and two new species of Pachygastrinae from the Oriental region (Diptera: Stratiomyidae). Zootaxa. 2402(1):61–67.
- Zhou QX, Ding SM, Li X, Zhang TT, Yang D. 2017. Complete mitochondrial genome of Allognosta vagans (Diptera, Stratiomyidae). Mitochondrial DNA B Resour. 2(2):461–462.