Abstract
The Macaroni penguin Eudyptes chrysolophus is a small crested penguin. In this study, the complete mitochondrial genome of E. chrysolophus is revealed for the first time. The mitogenome sequence is circular and 17,059 bp in length. It contains 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and two ribosomal RNA (rRNA) genes similar to other Spheniscidae species. The total nucleotide composition is 30.53% (A), 32.86% (C), 13.96% (G), and 22.66% (T), and 46.81% for overall GC contents. The phylogenetic analysis shows a close relationship between E. chrysolophus and E. schlegeli. Our findings would be useful for further studies on phylogenetics and evolutionary history of the genus Eudyptes.
The Macaroni penguin (Eudyptes chrysolophus), a species of crested penguin, is the most abundant penguin species worldwide, distributed from the Antarctic to the Subantarctic regions (Woehler and Poncet Citation1993). The population of macaroni penguin appears to have declined rapidly over the past three generations (BirdLife International Citation2020). The taxonomic status of the genus Eudyptes and the number of species within it have been repeatedly discussed (Frugone et al. Citation2018). Complete mitochondrial genomes provide baseline information to understand genomic evolution and facilitate phylogenetic inference (Matsui et al. Citation2009) and taxonomic clarifications (Sebastian et al. Citation2018). A mitogenomic study of E. chrysolophus have been reported; however, it was incomplete information (Cole et al. Citation2019). Thus, clarification of complete mitochondrial genome of E. chrysolophus is needed to understand their phylogeny and evolution.
Here, we sequenced and analyzed the complete mitogenome of E. chrysolophus (GenBank: MW074963). The blood sample (proof number MP1) was obtained at Narebski Point, Antarctica (62°14′14.66″S, 58°46′30.59ʺW), on 10 January 2010, and stored at the Korea Polar Research Institute (KOPRI), Incheon, South Korea. Total genomic DNA was extracted from the blood sample using a DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany). The complete mitochondrial genome was sequenced and annotated according to our previous study (Kim and Kim Citation2020). An Illumina paired-end (PE) library was prepared according to the manufacturer’s instructions with 550 bp inset size, and sequencing was performed using an Illumina sequencing platform supplied by a commercial company (Phyzen, Seongnam, South Korea).
The complete mitochondrial genome sequence of E. chrysolophus had a circular genome of 17,059 bp in length, containing 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and two ribosomal RNA (rRNA) genes. The overall base composition is 30.53% (A), 32.86% (C), 13.96% (G), and 22.66% (T). The heavy strand codes 28 genes and the light strand codes 9 genes. The 13 PCGs of E. chrysolophus encode 3798 amino acids. Most PCGs started with ATG codon, except for COI and ND3 which use the initiation codon GTG and ATT, respectively.
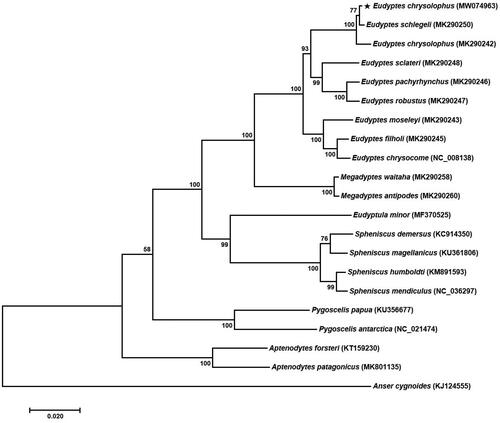
In order to ascertain the phylogenetic status of E. chrysolophus, a phylogenetic analysis was performed comparing the mitogenome of the E. chrysolophus with the inclusion of 21 published mitogenomes from the family Spheniscidae species. A phylogenetic tree was constructed by the maximum likelihood (ML) method with 1000 bootstrap replicates, using the MEGA 7.0 program (Kumar et al. Citation2016) and applying the Tamura–Nei model (). The phylogenetic relationship revealed that E. schlegeli (MK290250) was the most closely related species to E. chrysolophus (MW074963) as a sister group. Although E. chrysolophus (MK290242) is already registered at the NCBI as a partial sequence, the results of a phylogenetic analysis based only on PCGs for the same species showed slightly lower similarity compared with E. schlegeli (MK290250). We believe it would be worth studying inter-species mutations in further studies.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW074963. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA667576, SRR12778075, and SAMN16378289 respectively.
Additional information
Funding
References
- BirdLife International. 2020. Species factsheet: Eudyptes chrysolophus. [accessed 2020 Oct 13]. http://www.birdlife.org
- Cole TL, Kieren DT, Mitchell KJ, Tennyson AJD, Thomas DB, Pan H, Zhang G, Rawlence NJ, Wood JR, Bover P, Bouzat JL, et al. 2019. Mitogenomes uncover extinct penguin taxa and reveal island formation as a key driver of speciation. Mol Biol Evol. 36(4):784–797.
- Frugone MJ, Lowther A, Noll D, Ramos B, Pistorius P, Dantas GPM, Petry MV, Bonadonna F, Steinfurth A, Polanowski A, et al. 2018. Contrasting phylogeographic pattern among Eudyptes penguins around the Southern Ocean. Sci Rep. 8(1):17481
- Kim JU, Kim JH. 2020. Complete mitochondrial genome and phylogenetic analysis of the Weddell seal Leptonychotes weddellii. Mitochondrial DNA Part B. 5:3357–3358.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Matsui A, Rakotondraparany F, Munechika I, Hasegawa M, Horai S. 2009. Molecular phylogeny and evolution of prosimians based on complete sequences of mitochondrial DNAs. Gene. 441(1–2):53–66.
- Sebastian W, Sukumaran S, Zacharia PU, Gopalakrishnan A. 2018. The complete mitochondrial genome and phylogeny of Indian oil sardine, Sardinella longiceps and Goldstripe Sardinella, Sardinella gibbosa from the Indian Ocean. Conserv Genet Resour. 10(4):735–739.
- Woehler EJ, Poncet S. 1993. The distribution and abundance of Antarctic and Subantarctic Penguins. Cambridge: Scientific Committee on Antarctic Research.