Abstract
We report the first mitochondrial genome sequences for the three band pennant fish, Heniochus chrysostomus. The whole mitogenome of H. chrysostomus was circular in shape and 16,650 bp in length. The mitogenome consists of 13 typical vertebrate protein-coding genes, 22 tRNA genes, 2 rRNA genes (12S rRNA and 16S rRNA), and 2 putative non-coding regions. Phylogenetic tree analysis revealed that H. chrysostomus was closely related to Heniochus diphreutes. This study will provide useful genetic information for future phylogenetic and taxonomic classification of Chaetodontidae.
Heniochus chrysostomus (Cuvier, 1831) belongs to the Family Chaetodontidae and the Order Perciformes, and it looks similar to the pennant coralfish but can be distinguished by the yellow tip to the snout and three distinctive diagonal black bands on the body (Parmentier et al. Citation2011). The species is distributed in Western India to Pitcairn Islands, north to southern Japan, south to Rowley Shoals, the south China sea, southern Queensland, and New Caledonia (Chen and Zhang Citation2015). In this study, complete mitochondrial genome of H. chrysostomus was characterized, which would provide genomic data for taxonomic resolution, population genetic structure and phylogenetic relationship.
Specimen was collected from Sanya, Hainan Province, Republic of China (N16°25'26″, E110°46'06″) in October 2019 and deposited in the Key laboratory of Fishery Ecology and Environment, Guangdong Province, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China (voucher specimen number: HC-1). Total genomic DNA was extracted from the muscle tissue using the Aidlab Genomic DNA Extraction Kit following the manufacturer’s instructions (Aidlab Biotech, Beijing, China). The mitogenomes of H. chrysostomus was sequenced by next-generation sequencing (Illumina HisSeq 4000; Guangzhou JiRui Gene Technology Co. Ltd. China). Clean data without sequencing adapters were de novo assembled by the NOVOPlasty software (Dierckxsens et al. Citation2017). We compared the assembled genome with three confirmed sequences by PCR and Sanger sequencing methods to evaluate the single-base accuracy of the assembled mitochondrial genome. In this study, we obtained the complete mitochondrial genome of H. chrysostomus. Its mitochondrial genome is deposited in the GenBank under accession number MW136414. For a better understanding of genetic status and the evolutionary study, we focused on the genetic information contained in the complete mitochondrial genomes of the fish.
The complete mitogenome of H. chrysostomus was circular in shape and 16,650 bp in length with 16.24% G content, indicating an obvious anti-guanine bias commonly observed in other fishes (Cheng et al. Citation2012; Shan et al. Citation2016). The overall base composition was 28.64% of A, 25.47 of %T, 29.65% of C and 16.24% of G with a slight A + T bias (54.11%) like other vertebrate mitochondrial genomes. The mitogenome consists of 13 typical vertebrate protein-coding genes, 22 tRNA genes, 2 rRNA genes (12S rRNA and 16S rRNA), and 2 putative non-coding regions (control region and L-strand replication origin). All 13 protein-coding genes found in other vertebrates were also present in H. chrysostomus including three subunits of the cytochrome c oxidase (COI-III), seven subunits of the NADH ubiquinone oxidoreductase complex (ND1-6, ND4L), one subunit of the ubiquinol cytochrome oxidoreductase complex (Cyt b), and two subunit of ATP synthases (ATP6 and ATP8). The total length of those genes was 11,416 bp, accounting for 68.56%. Furthermore, as in other bony fishes, the mitogenome contained 22 tRNA genes interspersed between the rRNA and protein-coding genes. 14 tRNA genes were transcribed on the H-strand, whereas other 8 tRNA genes were oriented to the L-strand (Cheng et al. Citation2012; Li et al. Citation2013). Two of the 22 tRNA were determined for serine (UCN and AGY) and leucine (UUR and CUN), and one specific tRNA gene for the other amino acids. The tRNA genes varied in size from 65 bp to 75 bp.
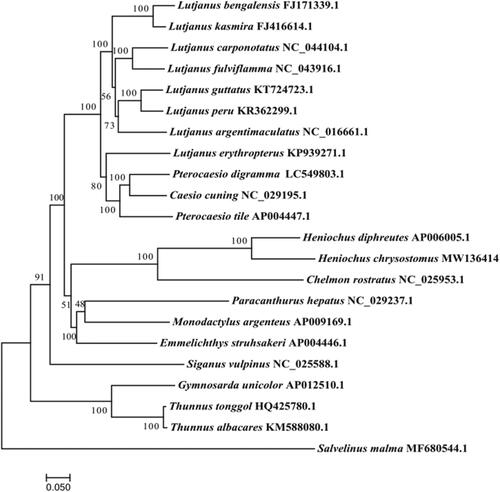
To determine the taxonomic status of H. chrysostomus, we constructed the phylogenetic tree using the maximum likelihood method on the basis of the complete sequences of the mitochondrial genomes of 21 species (Yang et al. Citation2017), and we selected Salvelinus malma as the outgroup. The phylogenetic tree showed that the H. chrysostomus had the closer relationship with Heniochus diphreutes (). The complete mitochondrial genome sequence of H. chrysostomus provided an important dataset for a better understanding of the mitogenomic diversities and evolution in fish as well as novel genetic markers for studying population genetics and species identification.
Figure 1. Phylogenetic tree of 22 complete mitogenomic sequences using the maximum likelihood method. Compared species: Lutjanus bengalensis, Lutjanus kasmira, Lutjanus carponotatus, Lutjanus fulviflamma, Lutjanus guttatus, Lutjanus peru, Lutjanus argentimaculatus, Lutjanus erythropterus, Pterocaesio digramma, Caesio cuning, Pterocaesio tile, Heniochus diphreutes, Chelmon rostratus, Paracanthurus hepatus, Monodactylus argenteus, Emmelichthys struhsakeri, Siganus vulpinus, Gymnosarda unicolor, Thunnus tonggol, Thunnus albacares.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in [National Center for Biotechnology Information] at [https://www.ncbi.nlm.nih.gov/], reference number [MW136414].
Additional information
Funding
References
- Chen DG, Zhang MZ. 2015. Marine fishes of China. Qingdao: Ocean University of China Press.
- Cheng J, Ma G, Song N, Gao T. 2012. Complete mitochondrial genome sequence of bighead croaker Collichthys niveatus (Perciformes, Sciaenidae): a mitogenomic perspective on the phylogenetic relationships of Pseudosciaeniae. Gene. 491(2):210–223.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Li N, Zhang Z, Zhao L, Gao T. 2013. Complete mitochondrial DNA sequence of the Pacific sand lance Ammodytes hexapterus (Perciformes: Ammodytidae): Mitogenomic perspective on the distinction of Ammodytes hexapterus and Ammodytes personatus. Mitochondrial DNA. 24(5):463–465.
- Parmentier E, Boyle K, Berten L, Christophe B, Lecchini D. 2011. Sound production and mechanism in Heniochus chrysostomus (Chaetodontidae). J Exp Biol. 214(Pt 16):2702–2708.
- Shan B, Song N, Han Z, Wang J, Gao T, Yokogawa K. 2016. Complete mitochondrial genomes of three sea basses Lateolabrax (Perciformes, Lateolabracidae) species: genome description and phylogenetic considerations. Biochem Syst Ecol. 67:44–52.
- Yang L, Meng F, Wang R, Shi G. 2017. Complete mitochondrial genome of the Salvelinus malma sp. (Salmoniformes, Salmonidae) with phylogenetic consideration. Mitochondrial DNA Part B. 2(2):889–890.
