Abstract
This study reports the complete mitochondrial genome of the Capsaloides cristatus (Monogenea: Capsalidae) collected from the gill lamella of Istiophorus platypterus. The total length of the mitogenome was 13,948 bp, containing 12 typical platyhelminthic protein-coding genes, 22 tRNA genes, 2 rRNA genes and a putative non-coding region, with the atp8 gene being absent. The total A + T content was 65.99%, which was significantly higher than that of the C + G content (34.01%). There were two kinds of start codons (ATG and GTG) and three kinds of terminated codons (TAA, TAG and TGA) in the 12 protein-coding genes. Phylogentic analysis revealed close relationships among the genera Capsaloides, Capsala, Benedenia and Neobenedenia with high bootstrap value supported. This study will provide useful molecular data for a better understanding of the species identification and phylogenetic position of C. cristatus.
The family Capsalidae Baird, 1853 comprises approximately 200 monogenean species, most of which are ectoparasites on marine fishes and some are important pathogens of cultivated fishes. The taxonomic identities of the members of the genus Capsaloides Price, 1938 are not easy to confirm due to deficient descriptions or illustrations (or both) of many of the nominal species (Chisholm & Whittington, Citation2006). Capsaloides cristatus Yamaguti, 1968 is a common external pathogen of the sailfish and sometimes lead to inflammation, mucus hyperproduction and hemorrhage of their hosts (Paperna, Citation1991). According to Chisholm & Whittington (Citation2006), C. cristatus can be distinguished with other species of the closely related species by the morphology of the haptoral accessory sclerites and the number and shape of dorsomarginal body sclerites. Though the general morphology is conserved, some morphological characters of C. cristatus can be easily influenced by various factors such as the parasitizing sites, the salinity and temperature in the environment, and the growth stage of parasite (Whittington et al., Citation2004). As a result, this study sequenced the complete mitochondrial genome of C. cristatus, aiming to provide useful molecular data for the identification and an improved understanding of the phylogenetic position of this parasite.
Specimens of C. cristatus were collected from the gill lamella of the Atlantic sailfish (Istiophorus platypterus) from Shanwei city, Guangdong Province, China (21°24′7.2″N, 115°19′51.6″E) in April 2019. The specimen was deposited in the Key Laboratory of South China Sea Fishery Resources Exploration & Utilization, Ministry of Agriculture and Rural Affairs, South China Sea Fisheries Research Institute, Chinese Academy of Fisheries Sciences under the voucher number CC20190912. By using the E.Z.N.A® Tissue DNA kit (OMEGA, USA), the total genomic DNA was extracted from the tissue of C. cristatus. The paired-end DNA library with insert size of 300−500 bp was constructed and sequenced by next generation sequencing (Illumina HisSeq 4000). The clean data without sequencing adapters were de novo assembled by NOVOplasty software (Loh et al., Citation2016). The assembled mitochondrial genes were identified by BLAST searches in the NCBI database. The locations of the protein-coding genes (PCGs) were determined using ORF Finder via NCBI, and the tRNA genes were verified using the MITOS WebServer (http://mitos2.bioinf.uni-leipzig.de/index.py). The start codons and stop codons of PCGs were identified based on comparisons with other monogeneans. Within the range of the previously reported monogenean mitogenomes, the whole mitochondrial genome of C. cristatus was 13,948 bp in length. The total A + T content (65.99%) was higher than the C + G content (34.01%) with a low G content (19.06), indicating an anti-guanine bias of the mitogenome. The mitogenome of C. cristatus consisted of 12 typical platyhelminthic PCGs, 22 tRNA genes, 2 rRNA genes and a putative non-coding region. According with the previously reported monogeneans (Plaisance et al., Citation2007; Huyse et al., Citation2008; Perkins et al., Citation2010; Kang et al., Citation2012; Zhang et al., Citation2014; Yang et al., Citation2020), all of the mitochondrial genes of C. cristatus were transcribed from the heavy strand, with the atp8 gene being absent.
All of the 12 PCGs found in other monogeneans were also present in C. cristatus, including one cytochrome b subunit (cytb), one ATP synthase subunit (atp6), three cytochrome c oxidase subunits (cox1-3), and seven NADH dehydrogenase subunits (nad1-6, nad4L). Three of the 12 protein-coding genes (nad1, nad2 and nad4L) started with the codon GTG, while the left nine genes (cytb, atp6, cox1-3 and nad3-6) used the start codon ATG. Interestingly, six of the 12 protein-coding genes (cox1, cytb, nad1, nad3, nadL and nad6) were inferred to end with the TAG terminated codon, four genes ended with the TAA terminated codon (nad2, nad4, nad5 and atp6), while the gene cox2 and cox3 ended with the codon TGA and TA-, respectively. Among the protein-coding genes, the longest one was cox1 with a length of 1572 bp, whereas the shortest one was nad4L (246 bp). The 22 tRNA genes folding into cloverleaf secondary structures were determined, with their sizes ranging from 58 bp to 71 bp. Separated by the trnC gene, the two rRNA genes rrnL and rrnS were located between cox2 and trnT, with a length of 948 bp and 736 bp, respectively. In addition, the none-coding region was confirmed to be 735 bp in length, and was determined to locate between the trnV and trnQ gene. The A + T content (79.05%) of the non-coding region was significantly higher than that of the overall mitochondrial genome (65.99%). This result was similar to those previously reported by Kang et al. (Citation2012) for Benedenia hoshinai, Zhang et al. (Citation2014) for Neobenedenia melleni and Yang et al. (Citation2020) for Capsala pricei.
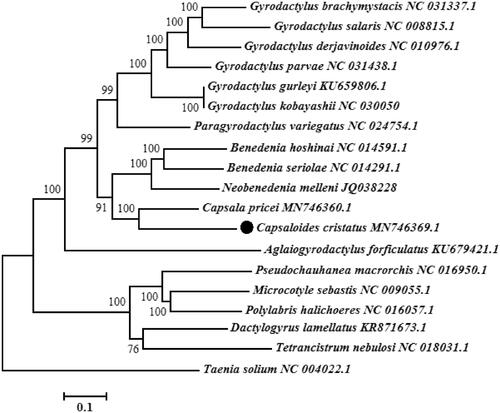
To analyze the phylogenetic position of C. cristatus, we constructed a phylogenetic tree using the maximum likelihood (1000 bootstrap replicates) method on the basis of 12 PCGs of 17 monogenean species. For contrasting the tree topology, Taenia solium (Cestoda) was selected as outgroup (Yang et al., Citation2020). The phylogenetic tree showed that C. cristatus was clustered with C. pricei, B. seriolae, B. hoshinai and N. melleni (), suggesting close relationships among the genera Capsaloides, Capsala, Benedenia and Neobenedenia. The complete mitochondrial genome sequence of C. cristatus provided important dataset for a better understanding of the species identification and mitogenomic evolution of monogeneans.
Acknowledgements
We thank the editor and anonymous reviewers for their valuable comments on the manuscript.
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MN746369.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA592303, SRS5732812, and SAMN13420506, respectively.
Additional information
Funding
References
- Chisholm LA, Whittington ID. 2006. Revision of Capsaloides (Monogenea: Capsalidae) with a redescription of C. magnaspinosus Price, 1939 from the nasal tissue of Tetrapterus audax (Istiophoridae) collected off Nelson Bay, New South Wales. Australia. Zootaxa. 1160(1):1–20.
- Huyse T, Buchmann K, Littlewood DTJ. 2008. The mitochondrial genome of Gyrodactylus derjavinoides (Platyhelminthes: Monogenea)-a mitogenomic approach for Gyrodactylus species and strain identification. Gene. 417(1-2):27–34.
- Kang S, Kim J, Lee J, Kim S, Min G, Park J. 2012. The complete mitochondrial genome of an ectoparasitic monopisthocotylean fluke Benedenia hoshinai (Monogenea: Platyhelminthes). Mitochondrial DNA. 23(3):176–178.
- Loh KH, Shao KT, Chen HM, Chen CH, Chong VC, Loo PL, Shen KN, Hsiao CD. 2016. Next generation sequencing yields the complete mitochondrial genome of the Zebra moray. Gymnomuraena zebra (Anguilliformes: Muraenidae). Mitochondrial DNA. 27(6):4230–4231.
- Paperna I. 1991. Diseases caused by parasites in the aquaculture of warm water fish. Ann Rev Fish Dis. 1:155–194.
- Perkins EM, Donnellan SC, Bertozzi T, Whittington ID. 2010. Closing the mitochondrial circle on paraphyly of the Monogenea (Platyhelminthes) infers evolution in the diet of parasitic flatworms. Int J Parasitol. 40(11):1237–1245.
- Plaisance L, Huyse T, Littlewood DTJ, Bakke TA, Bachmann L. 2007. The complete mitochondrial DNA sequence of the monogenean Gyrodactylus thymalli (Platyhelminthes: Monogenea), a parasite of grayling (Thymallus thymallus). Mol Biochem Parasitol. 154(2):190–194.
- Whittington ID, Deveney MR, Morgan JAT, Chisholm LA, Adlard RD. 2004. A preliminary phylogenetic analysis of the Capsalidae (Platyhelminthes: monogenea: Monopisthocotylea) inferred from large subunit rDNA sequences. Parasitology. 128(Pt 5):511–519.
- Yang CP, Shan BB, Liu Y, Zhao Y, Sun DR. 2020. Next generation sequencing yields the complete mitochondrial genome of the Capsala pricei Hidalgo, 1959 (Platyhelminthes: Monogenea) from South China Sea. Mitochondrial DNA Part B. 5(2):1964–1966.
- Zhang J, Wu XY, Li YW, Zhao MW, Xie MQ, Li AX. 2014. The complete mitochondrial genome of Neobenedenia melleni (Platyhelminthes: Monogenea): mitochondrial gene content, arrangement and composition compared with two Benedenia species. Mol Biol Rep. 41(10):6583–6589.