Abstract
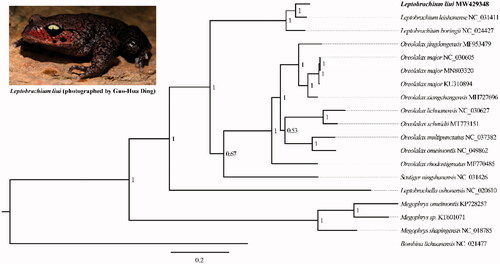
The Chong’an Mustache Toad, Leptobrachium liui (Pope, 1947) is a Chinese endemic species, inhabiting the mountain streams with rich vegetation in southeastern China. The first complete mitochondrial genome (mitogenome) of L. liui was assembled using the data of whole-genome sequencing. The size of the complete mitogenome for L. liui was 17,190 bp, which included 13 PCGs, 23 tRNAs with two concatenated tRNAMet genes, 2 rRNAs, a non-coding region, and a D-loop. The Bayesian tree shows that L. liui was positioned near L. leishanense within the genus Leptobrachium.
The genus of Leptobrachium Tschudi, 1838 currently contained 37 species distributed in Southern China and India to islands of the Sunda Shelf and the Philippines (Frost Citation2020). Previously, five Leptobrachium species with spines on the upper lip in adult males were classified as Vibrissaphora Liu, 1945 (Fei and Ye Citation2016), but phylogeny showed that Vibrissaphora was not a subgenus and placed within the genus of Leptobrachium (Matsui et al. Citation2010). At present, only the complete mitochondrial genomes (mitogenomes) of Leptobrachium leishanensis (Liang et al. Citation2016) and Leptobrachium boringii (Li et al. Citation2019) could be download from the Genbank database. Therefore, more complete mitogenomes of this genus need to be obtained for phylogenetic studies. The Chong’an Mustache Toad, Leptobrachium liui (Pope, 1947) is a Chinese endemic species, which mainly inhabits in the mountain streams with rich vegetation in southeastern China (Fujian, Jiangxi, and Zhejiang) (Frost Citation2020). Here, we firstly reported the complete mitogenome of L. liui, representing the third mitogenome from the genus Leptobrachium.
The specimens of L. liui were captured from the Jiulongshan National Nature Reserve (N28.371988°, E1118.898758°) in Suichang County, Zhejiang Province, eastern China. Then these samples were preserved in 90% ethanol at the Museum of Laboratory of Amphibian Diversity Investigation (contact person: Guo-Hua Ding, E-mail: [email protected]), Lishui University, China. We randomly selected an adult sample (species voucher: LSU20191108JLS001) with the ethanol removed and finished the DNA extraction using the EasyPure genomic DNA kit (TransGen Biotech Co, Beijing, China). Preparation and sequencing of the whole genomic DNA library were finished in Novogene Bioinformatics Technology Co. Ltd. (Tianjin, China). Genomic DNA was fragmented with Covaris to an average insert size of 350 bp and sequenced on the Illumina NovaSeq 6000 platform for paired-end 150 bp. By using NOVO Plasty 3.7 (Dierckxsens et al. Citation2017), we assembled the mitogenome of L. liui from the data of whole-genome sequencing. Then The position and direction of 13 protein-coding genes (PCGs), 2 rRNA (16S and 12S) genes, 23 tRNA genes, a non-coding region (NCR) of an L-strand replication origin, and a D-loop were predicted by MITOS WebServer (Matthias et al. Citation2013) and tRNA-scan (Chan and Lowe Citation2019).
The length of complete mitogenome of L. liui was 17,190 bp (Genbank accession: MW429348) with 28.1% A, 32.6% T, 14.9% G, and 24.4% C, the higher value of A + T content (60.7%) compare to G + C content (39.3%), indicating there was slight A + T bias in L. liui. The arrangement pattern and transcription directions of mitogenome of L. liui were identical to previous studies in the genus Leptobrachium (Liang et al. Citation2016; Li et al. Citation2019). Two tRNAMet genes (tRNAMet1 and tRNAMet2) were derived from tandem duplication in this mitogenome. The D-loop (1492 bp) was found between tRNATrp and tRNAPhe, and the NCR (28 bp) is found between tRNAAsn and tRNACys. The total length of the 13 PCGs was 11,394 bp. Nine of them (ND1, ND2, COII, ATP6, COIII, ND4L, ND5, ND6, and CYTB) started with ATG as a start codon, while COI, ATP8, ND3, and ND4 started with GTG. Two PCGs (ND5 and ND6) end with AGG, four PCGs (ND2, COI, ATP8, and ND4) used completed stop codon TAA, whereas the other seven PCGs ended with an incompleted stop codon (T in COII, COIII, ND3, ND4, and CYTB and TA in ND1 and ATP6).
Bayesian inference tree was constructed with concatenated 13 PCGs on MrBayes v3.2.2 setting 4 independent chains and executing 1,000,000 generations based on the published complete mitogenome sequences of the family Megophryidae. Bombina lichuanensis (Anura: Bombinatoridae) was chosen as an outgroup. As shown in . liui was positioned near L. leishanense within the genus Leptobrachium. The p-distances of the CYTB gene between L. liui and the other two Leptobrachium species were both more than 7.98% by MEGA 5.05. In addition, the genus Megophrys was a basal clade relative to others within The family Megophryidae, which is similar to previous results (Pyron and Wiens Citation2011; Liang et al. Citation2016). The complete mitogenome of L. liui in this study will be an important genetic resource to the studies of conservation and restoration of L. liui. Furthermore, it will play an important role in understanding the molecular evolution of the family Megophryidae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The mitogenome data supporting this study are openly available in GenBank at (https://www.ncbi.nlm.nih.gov/nuccore/MW429348). Reference number (Accession number: MW429348). BioSample and SRA accession numbers are (https://www.ncbi.nlm.nih.gov/biosample/SAMN17205596), (https://www.ncbi.nlm.nih.gov/sra/SRR13345317), respectively.
Additional information
Funding
References
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Fei L, Ye CY. 2016. Amphibians of China. Vol. I. Beijing (China): Science Press; p. 466–485.
- Frost DR. 2020. Amphibian species of the world: an online reference, version 6.0. New York (NY): American Museum of Natural History. [accessed 2021 January 4]. http://research.amnh.org/herpetology/amphibia/index.php/
- Li YW, Li C, Chen QL, Liu ZH, Shen YJ. 2019. The mitochondrial genome for tadpole of Emei moustache toad (Leptobrachium boringii: Anura: Megophryidae) from the Southwest China and its phylogenetic analysis. Mitochondrial DNA B. 4(1):265–266.
- Liang XX, Shu GC, Wang B, Jiang JP, Li C, Xie F. 2016. Complete mitochondrial genome of the Leishan moustache toad, Vibrissaphora leishanensis (Anura: Megophryidae). Mitochondrial DNA B Resour. 1(1):275–276.
- Matsui M, Hamidy A, Murphy RW, Khonsue W, Yambun P, Shimada T, Ahmad N, Belabut DM, Jiang JP. 2010. Phylogenetic relationships of megophryid frogs of the genus Leptobrachium (Amphibia, Anura) as revealed by mtDNA gene sequences. Mol Phylogenet Evol. 56(1):259–272.
- Matthias B, Alexander D, Frank J, Fabian E, Catherine F, Guido F, Joern P, Martin M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Pyron RA, Wiens JJ. 2011. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of advanced frogs, salamanders, and caecilians. Mol Phylogenet Evol. 61(2):543–583.