Abstract
In this study, we report the complete mitochondrial genomes (mtgenome) of Thyreus decorus, Ceratina okinawana and Amegilla calceifera, which are the first time of mtgenome report also for the genera Thyreus, Ceratina and Amegilla in the family Apidae. They contain 15,237, 15,207, and 17,728 bp, with AT content of 84.97%, 79.30%, and 84.63%, respectively. Each mtgenome includes 13 protein-coding genes (PCGs), 22 transfer RNA (tRNAs), two ribosomal RNA (16S and 12S rRNA) and an AT-rich control region (CR). The phylogenetic relationships of 45 species in the family were constructed using Bayesian Inference based on concatenated nucleotide sequences of 13 PCGs. Our study suggests that the subfamily Apinae is a paraphyletic group, with the genus Eucera claded into the subfamily Xylocopinae and the genera Amegilla and Thyreus into the subfamily Nomadinae. In Apinae, the genera Melipona and Bombus are significantly sister group, and the genus Apis is the sister group with Melipona + Bombus.
The family Apidae has over known 5900 species, the largest family in the superfamily Apoidea in the number of species, is classified into three subfamilies (Apinae, Nomadinae, and Xylocopinae) and 210 genera (Ascher and Pickering Citation2020). Species of the family are considered to be the main pollinators of angiosperm, and have an indispensable influence in the normal operation of natural habitats and agricultural ecosystems (Williams Citation1994; Pywell et al. Citation2006; Ollerton et al. Citation2011). In addition, some species of the family play an important role in the economy and are used for the commercial production of honey and pollination of cultivated crops (Del Sarto et al. Citation2005). The phylogenetic relationships have largely unsettled in the family (Hedtke et al. Citation2013; Bossert et al. Citation2019). In this study, we sequenced and annotated the complete mtgenomes of Thyreus decorus, Amegilla calceifera and Ceratina okinawana also for the first time for their belonging genera in Apidae. We constructed and discussed the phylogenetic relationships of 45 known mtgenomes in Apidae.
The samples of T. decorus, C. okinawana and A. calceifera used for this study were collected from Guangxi (21°90.527 N, 107°90.365E), Guangxi (21°90.537 N, 107°90.448E) and Chongqing (28°90.881 N, 106°33.622E), China, respectively. Individual specimens were preserved in absolute ethanol after morphological identification and then stored at −80 °C before the DNA extraction. The total DNA was extracted from the muscle tissues using the DNeasy Blood and Tissue kit (250) (Qiagen 69506, Duesseldorf, Germany), and deposited in the Institute of Entomology and Molecular Biology, Chongqing Normal University, China (accession numbers: WZQ-TD-1, WZQ-CO-1 and WZQ-AC-1). The complete mitochondrial genomes of the three species were sequenced using the Illumina HiSeq system in the Shenzhen Huitong Biotechnology Limited Company (Shenzhen, China), and annotated using Mitos (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013). The PCGs and rRNAs genes were compared with the mtgenomes of related species, and the annotation results were confirmed using Geneious v4.8.5 (Kearse et al. Citation2012).
The complete mtgenomes of T. decorus, C. okinawana and A. calceifera are 15,237, 15,207 and 17,728 bp in length. They have been submitted to GenBank with accession numbers MW281318∼MH281320. Each mtgenome comprises of 37 typical mtgenome genes (13 PCGs, 22 tRNAs, and two rRNAs) and a control region (CR), consistent with other species in Apidae (Zhao et al. Citation2017). The mtgenomes of the three species have a high nucleotide bias with 84.97%, 79.30%, and 84.63% A + T content, and 15.03%, 20.70%, and 15.37% G + C content, respectively.
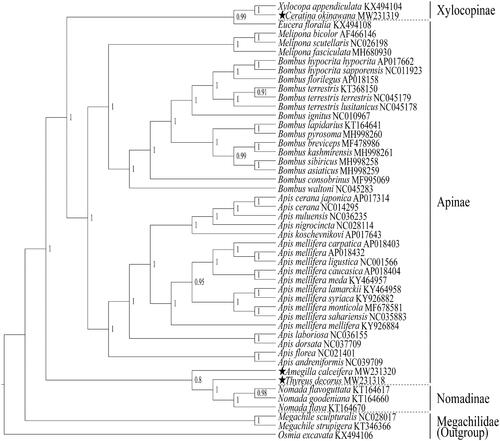
The phylogenetic relationships were constructed with the three newly sequenced and 42 known mtgenome sequences in Apidae, and Megachile sculpturalis, Megachile strupigera and Osmia excavata (Hymenoptera: Megachilidae) were used as outgroup. The 45 known species of mtgenome sequences were downloaded from GenBank. The Bayesian inference (BI) was employed for phylogenetic tree construction based on concatenated nucleotide sequences of 13 PCGs using PhyloSuite 1.2.1, and the best model was selected using ParririonFinder2 in the phylogenetic analysis (Ronquist et al. Citation2012; Zhang et al. Citation2020) (). The result suggests that the subfamily Apinae is a paraphyletic group, with the genus Eucera clustered into the subfamily Xylocopinae and the genera Amegilla and Thyreus claded into the subfamily Nomadinae. In Apinae, the three genera Melipona, Bombus and Apis are obviously monophyletic with one of posterior probability for all of the three general clades. The genera Melipona and Bombus are sister group, and the genus Apis is the sister group with Melipona + Bombus. The genus Eucera, traditionally classified as Apinae, was grouped into Xylocopinae clade with 0.99 of posterior probability, and it maybe should be classified into the subfamily Xylocopinae, which is consistent with earlier studies (Zheng et al. Citation2018). Similarly, and the genera Amegilla and Thyreue, both traditionally classified to Apinae, were grouped into Nomadinae clade with 0.8 of posterior probability, and they maybe should be classified into the subfamily Nomadinae. Our results are basically consistent with the results of the earlier phylogenetic research results (Kawakita et al. Citation2008; Danforth et al. Citation2013). However, the genera Eucera, Ceratina, Amegilla and Thyreue have each only one mtgenome sequence to be included in the phylogenetic analysis; therefore, their phylogenetic positions need a further argument with more mtgenome sequences involved.
Figure 1. The phylogenetic relationships are constructed using Bayesian inference based on nucleotide sequences of 13 PCGs with 45 species of the family Apidae, and three species in the family Megachilidae are used as outgroup. The Bayesian posterior probabilities are showed at the nodes. The three newly sequenced mtgenomes are indicated by pentagrams.
Disclosure statement
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW281318-MW281320. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA706124, PRJNA706149 and PRJNA706161, SRS8360571, SRS8361332 and SRS8361675, and SAMN18118499, SAMN18118864 and SAMN18119826 respectively.
Additional information
Funding
References
- Ascher JS, Pickering J. 2020. Discover life bee species guide and world checklist (Hymenoptera: Apoidea: Anthophila) https://www.discoverlife.org/mp/20q?guide=Apoidea_species&flags=HAS.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet E. 69(2):313–319.
- Bossert S, Murray EA, Almeida EAB, Brady SG, Blaimer BB, Danforth BN. 2019. Combining transcriptomes and ultraconserved elements to illuminate the phylogeny of Apidae. Mol Phylogenet E. 130:121–131.
- Danforth BN, Cardinal S, Praz C, Almeida EAB, Michez D. 2013. The impact of molecular data on our understanding of bee phylogeny and evolution. Annu Rev Entomol. 58:57–78.
- Del Sarto MCL, Peruquetti RC, Campos LAO. 2005. Evaluation of the neotropical stingless bee Melipona quadrifasciata (Hymenoptera: Apidae) as pollinator of greenhouse tomatoes. J Econ Entomol. 98(2):260–266.
- Hedtke SM, Patiny S, Danforth BN. 2013. The bee tree of life: A supermatrix approach to apoid phylogeny and biogeography. BMC Evol Biol. 13(1):138.
- Kawakita A, Ascher JS, Sota T, Kato M, Roubik DW. 2008. Phylogenetic analysis of the corbiculate bee tribes based on 12 nuclear protein-coding genes (Hymenoptera: Apoidea: Apidae). Apidologie. 39(1):163–175.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Ollerton J, Winfree R, Tarrant S. 2011. How many flowering plants are pollinated by animals? Oikos. 120(3):321–326.
- Pywell RF, Warman EA, Hulmes L, Hulmes S, Nuttall P, Sparks TH, Critchley CNR, Sherwood A. 2006. Effectiveness of new agri-environment schemes in providing foraging resources for bumblebees in intensively farmed landscapes. Biol Conserv. 129(2):192–206.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. Mrbayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Williams IH. 1994. The dependences of crop production within the European Union on pollination by honey bees. Agric Zool Rev. 6:229–257.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
- Zhao X, Wu Z, Huang J, Liang C, An J, Sun C. 2017. Complete mitochondrial genome of Bombus breviceps (Hymenoptera: Apidae). Mitochondrial DNA B Resour. 2(2):604–606.
- Zheng BY, Cao LJ, Tang P, van Achterberg K, Hoffmann AA, Chen HY, Chen XX, Wei SJ. 2018. Gene arrangement and sequence of mitochondrial genomes yield insights into the phylogeny and evolution of bees and sphecid wasps (Hymenoptera: Apoidea). Mol Phylogenet Evol. 124:1–9.
