Abstract
Melicope pteleifolia commonly known as thin evodia, is an herb used to therapy eczema, dermatitis, and other ailments in traditional Chinese medicine. Here, we reported the third complete chloroplast genome of M. pteleifolia based on next-generation sequencing. The third chloroplast genome of M. pteleifolia is 158,933 bp in length consisting of large and small single-copy regions of length 85,020 and 18,607 bp, separated by two IR regions of 27,683 bp. The overall GC content was 38.30%. De novo assembly and annotation showed the chloroplast genome of M. pteleifolia encodes 134 genes, including 89 protein-coding genes, 37 tRNA genes, and eight rRNA genes. A huge intraspecies variation was found with 248 SNPs and 97 INDELs among three assemblies of M. pteleifolia. Phylogenetic tree indicated that three assemblies of M. pteleifolia form a clade, sister to the genus Phellodendron and Casimiroa.
Melicope pteleifolia (Rutaceae) is a shrub commonly distributed in south of China and used as a traditional Chinese medicine to treat eczema, dermatitis, rheumatic arthralgia, and other ailments (Flora of China Committee Citation2008; Yao et al. Citation2020). Modern pharmacological studies reported that its crude extracts exhibited analgesic, anti-inflammatory, anti-tumor, and antioxidative effects (Shaari et al. Citation2011; Nguyen et al. Citation2016; Kabir et al. Citation2018; Lee et al. Citation2019). So, it is worth doing some works to utilize M. pteleifolia better, including distinct M. pteleifolia from its closely related species and investigate its intraspecies variation to ensure the safety of usage. However, very less is known about the genomics information of M. pteleifolia, even the genus Melicope. Up to now, chloroplast genomes from about 26 species of Rutaceae have been sequenced and published, and two of them belongs to the M. pteleifolia. To evaluate the intraspecies variation, we sequenced the third chloroplast genome of a M. pteleifolia individual growing in wild field in the Jingxi, Guangxi province (105°58′E, 23°06′N), 780 km away from the first individual isolated (Yu et al. Citation2021). The relationship between M. pteleifolia and other Rutaceae species was analyzed in this article with hope to provide better understanding of the phylogenetic status of Melicope and M. pteleifolia.
We collected fresh healthy leaves from M. pteleifolia species growing in the Jingxid, Guangxi province (105°58′E, 23°06′N). Voucher specimen was stored in herbarium of Institute of Chinese Materia Medica (CMMI, accession number 451025LY0636), China Academy of Chinese Medical Sciences. The DNA extraction and sequencing were performed as described before (Liu et al. Citation2020). Briefly, the sequencing library was constructed using NEB Next® Ultra DNA Library Prep Kit for Illumina® (NEB, Ipswich, MA). Paired-end (2 × 150 bp) sequencing was performed by Novogene Bioinformatics Technology Co. Ltd (Beijing, China), using the Illumina Hiseq X-Ten platform. About 5.0 Gb of sequence data was obtained after sequencing and base quality control. The paired-end reads were then assembled with GetOrganelle (Jin et al. Citation2020) based on the default reference sequences. The complete genome sequence was annotated by both GeSeq (Tillich et al. Citation2017) and PGA (Qu et al. Citation2019) based on previously reported Amborella trichopoda chloroplast genome (NC_005086.1) and Citrus reticulata chloroplast genome (NC_034671.1). Finally, we checked and merged the annotation from GeSeq and PGA manually. The annotated genomic sequence had been submitted to GenBank with the accession number MW263046.
The chloroplast genome of M. pteleifolia is 158,933 bp in length consisting of large and small single-copy regions of length 85,020 and 18,607 bp, separated by two IR regions of 27,683 bp. GC content was 38.30%. The genome consisted of 134 different genes, including 89 protein-coding genes, 37 distinct tRNA genes, and eight rRNA genes.
Based on the alignment of the three chloroplasts of M. pteleifolia, there is significant difference between our sequence and the published sequences. 247 single nucleotide polymorphisms (SNPs) and 96 insertions and deletions (INDELs) were identified between our sequence and MW046256. 121 SNPs and 26 INDELs were found in CDS region. Number of intraspecific variations identified between our sequence and NC_050882 is large compared to variations between NC_050882 and MW046256 (4 SNPs and 7 INDELs) or other species (Park et al. Citation2019; Park and Oh Citation2020), indicating high-level genetic diversity exists in M. pteleifolia. Results in this study suggested that the previous samples (NC_050882 and MW046256) may come from the same population, but our sample collected in Guangxi may come from a different population.
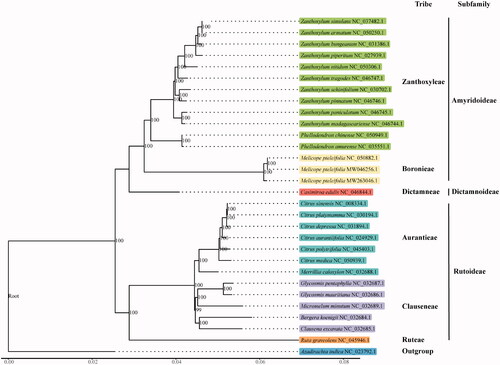
To confirm the phylogenetic location of M. pteleifolia within the family of Rutaceae, a total of 28 complete cp genomes of Rutaceae (including two previous M. pteleifolia chloroplast assemblies) were obtained from GenBank, and Azadirachta indica in Meliaceae family was used as out-group. The 30 complete chloroplast sequences were aligned by the MAFFT version 7 software (Katoh and Standley Citation2013). Phylogenetic analysis was conducted based on maximum likelihood (ML) analyses implemented in IQ-TREE version 2.0.5 (Minh et al. Citation2020) under the TVM + F + R2 nucleotide substitution model, which was selected by ModelFinder (Kalyaanamoorthy et al. Citation2017). Support for the inferred ML tree was inferred by bootstrapping with 1000 replicates. Phylogenetic analysis results strongly supported that M. pteleifolia was closely related to the genus Phellodendron and Casimiroa ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW263046.
Additional information
Funding
References
- Flora of China Committee. 2008. Flora of China. Beijing, China: Science Press.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li D-Z. 2020. Getorganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Kabir MF, Mohd Ali J, Haji Hashim O. 2018. Microarray gene expression profiling in colorectal (hct116) and hepatocellular (hepg2) carcinoma cell lines treated with Melicope ptelefolia leaf extract reveals transcriptome profiles exhibiting anticancer activity. PeerJ. 6:e5203.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. Modelfinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. Mafft multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lee BW, Park JG, Ha TKQ, Pham HTT, An JP, Noh JR, Lee CH, Oh WK. 2019. Constituents of the edible leaves of Melicope pteleifolia with potential analgesic activity. J Nat Prod. 82(8):2201–2210.
- Liu H, Zhao Y, Zhou J, Ma Q, Wang X, Hua Z. 2020. Complete chloroplast genome sequence of Murraya paniculata (rutaceae): a widely used folk medicinal herb. Mitochondrial DNA Part B. 5(3):3696–3697.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. Iq-tree 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Nguyen NH, Ha TK, Choi S, Eum S, Lee CH, Bach TT, Chinh VT, Oh WK. 2016. Chemical constituents from Melicope pteleifolia leaves. Phytochemistry. 130:291–300.
- Park J, Kim Y, Kwon W, Nam S, Song MJ. 2019. The second complete chloroplast genome sequence of Nymphaea alba L. (Nymphaeaceae) to investigate inner-species variations. Mitochondrial DNA Part B. 4(1):1014–1015.
- Park J, Oh SH. 2020. A second complete chloroplast genome sequence of Fagus multinervis Nakai (Fagaceae): intraspecific variations on chloroplast genome. Mitochondrial DNA Part B. 5(2):1868–1869.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Shaari K, Suppaiah V, Wai LK, Stanslas J, Tejo BA, Israf DA, Abas F, Ismail IS, Shuaib NH, Zareen S, et al. 2011. Bioassay-guided identification of an anti-inflammatory prenylated acylphloroglucinol from melicope ptelefolia and molecular insights into its interaction with 5-lipoxygenase. Bioorg Med Chem. 19(21):6340–6347.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. Geseq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Yao Q, Gao Y, Lai C, Wu C, Zhao CL, Wu JL, Tang DX. 2020. The phytochemistry, pharmacology and applications of Melicope pteleifolia: a review. J Ethnopharmacol. 251:112546.
- Yu J, Chen L, Mao J, Jin X, Shen J. 2021. The complete chloroplast genome of Melicope Pteleifolia (Rutaceae), a traditional medicinal plant in southeast China. Mitochondrial DNA B Resour. 6(1):60–61.