Abstract
The mitogenome data of leafhopper species Mileewa alara was assembled and annotated in this study. The results shows that length of M. alara is 16020 bp, consist of 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and one control region. The A + T content in the mitogenome was 77.9%. Phylogenetic analysis based on 13 PCGs of four Mileewa species and other 29 Cicadellidae species, each subfamily species well separated. And M. alara clustered with M. ponta. This study also raised mitogenome of Mileewa number in GenBank to four.
Mileewa Distant (Citation1908), is a leafhopper genus that belongs to tribe Mileewini of subfamily Mileewinae based on research Dietrich (Citation2011), that inhabit montane cloud forests where they occur on herbaceous vegetation in the understory. The Mileewa genus now comprises 49 species and well described in China (Yang et al. Citation2017). In this study, we sequenced and analyzed M. alara mitochondrial genome in order to provide more molecular evidence of genus Mileewa phylogeny.
We used two male adults of M. alara which were collected at Diaoluo mountain, Hainan, China (109°52′42″E, 18°43′7″N) on 17, June, 2019. Genomic DNA was extracted from the entire body without abdomen and wings by using Qiagen DNeasy Blood and Tissue Kit following manufacturer’s instructions. The mitochondrial genome of M. alara was sequenced by Illumina NovaSeq6000 platform (Berry Genomics, Beijing, China). Total 7.43 Gbp sequence raw reads were assembled by NOVOPlasty v4.0 (Dierckxsens et al. Citation2017) then annotated by using MitoZ v2.3 (Meng et al. Citation2019) and MITOS2 webserver (Bernt et al. Citation2013). All tRNA genes were identified by ARWEN v1.2 (Laslett and Canbäck Citation2008). The mitogenome data of M. alara was submitted to GenBank with accession number MW533151. The male genitalia were deposited at the Institute of Entomology, Guizhou University, Guiyang, China (GUGC), with the vocher number GUGC-IDT-00522.
The mitogenome of M. alara is 16020 bp in length and included 37 typical genes. The A + T content is 77.9%, same as other Mileewa species (He et al. Citation2019, He and Yang Citation2020). Most of PCGs have ATN as start codon while ATP8 genes initiated with TTG, and great majority of them use TAA as stop codon, except COX2, which stop with an incomplete condon (T). The size of ribosomal RNA gene 16S rRNA and 12S rRNA are 1224 bp and 813 bp. All tRNAs is range from 62 bp (tRNA-I) to 72 bp (tRNA-K).
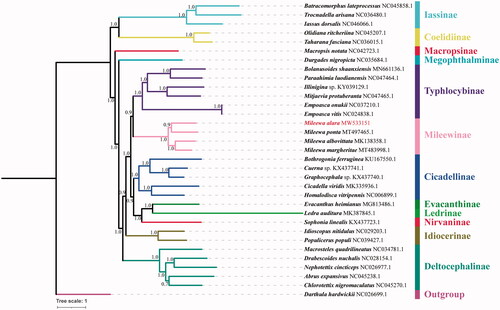
We downloaded 32 species mitochondrial genomes from GenBank which used for phylogenetic analysis with M. alara. The sequences of all 13 PCGs genes for each of the above species were extracted from GenBank files using PhyloSuite v1.2.2 (Zhang et al. Citation2020), then 13 sequences were aligned in batches with MAFFT v7.313 (Katoh and Standley Citation2013) using ‘–auto’ strategy and codon alignment mode. The alignments were refined using the codon-aware program MACSE v. 2.03 (Ranwez et al. Citation2018). Ambiguously aligned fragments of 13 alignments were removed in batches using Gblocks (Talavera and Castresana Citation2007). ModelFinder (Kalyaanamoorthy et al. Citation2017) was used to select the best-fit partition model (Edge-linked) using BIC criterion. Bayesian phylogenies were inferred using MrBayes 3.2.6 (Ronquist et al. Citation2012) under partition model (2 parallel runs, 303200 generations), in which the initial 25% of sampled data were discarded as burn-in. The tree showed that genus Mileewa species were clustered in one well supported clade ((M. ponta + M. alara) + (M. margheritae + M. albovittata)) and have sister relationships with Typhlocybinae species (). The phylogenetic results were very similar to our previous study (He et al. Citation2019, He and Yang Citation2020). Meanwhile, we still need more different genus mitogenome sequence data to rebuild Mileewinae phylogenetic relationships.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in [NCBI] at [https://www.ncbi.nlm.nih.gov/], reference number [NC006899.1, MK335936.1, KX437741.1, KX437740.1, KU167550.1, NC045207.1, NC036015.1, NC045270.1, NC045238.1, NC034781.1, NC028154.1, NC026977.1, MG813486.1, NC046066.1, NC045858.1, NC036480.1, NC039427.1, NC029203.1, NC042723.1, NC035684.1, MK138358.1, KX437723.1, NC047465.1, NC047464.1, NC037210.1, NC024838.1, MT497465.1, MN661136.1, KY039129.1, MK387845.1, NC026699.1, MT483998.1, MW533151].
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: denovo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Dietrich CH. 2011. Tungurahualini, a new tribe of Neotropical leafhoppers, with notes on the subfamily Mileewinae (Hemiptera, Cicadellidae). ZooKeys. 124:19–39.
- Distant WL. 1908. The Fauna of British India including Ceylon and Burma. Rhynchota IV Homoptera and Appendix (Pt.). London: Taylor & Francis; p. 501.
- He HL, Li HX, Yang MF. 2019. Complete mitochondrial genome sequence of Mileewa albovittata (Hemiptera: Cicadellidae: Mileewinae). Mitochondrial DNA Part B Resour. 4(1):740–741.
- He HL, Yang MF. 2020. Characterization and phylogenetic analysis of the mitochondrial genome of Mileewa ponta (Hemiptera: Cicadellidae: Mileewinae). Mitochondrial DNA B Resour. 5(3):2994–2995.
- He HL, Yang MF. 2020. The mitogenome of Mileewa margheritae (Hemiptera: Cicadellidae: Mileewinae). Mitochondrial DNA Part B Resour. 5(3):3181–3182.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. Mitoz: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Ranwez V, Douzery EJP, Cambon C, Chantret N, Delsuc F. 2018. MACSE v2: toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol Biol Evol. 35(10):2582–2584.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577.
- Yang MF, Meng ZH, Li ZZ. 2017. Hemiptera: Cicadellidae (II): Cicadellinae. Fauna Sinica: Insecta, vol. 67. Beijing, China: Science Press.
- Zhang D, Gao F, Jakovlic I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.