Abstract
Sweet chestnut (Castanea sativa) is an endemic species of genus Castanea in Europe, which is widespread in the southern part of continental Europe. The complete genome sequence of chloroplast was determined through Illumina NovaSeq platform. Totally the genome of chloroplast was 160,938 bp in length, GC rich (36.8%), comprising a pair of 25,726 bp inverted repeat sequences, separated by a 90,519 bp large and 18,967 bp small single-copy regions. There were 129 genes, including 37 transfer RNA genes, 8 ribosomal RNA genes, and 84 protein-coding genes. The phylogenetic tree analysis showed that C. sativa exhibited the closest relationship with Castanea henryi.
Sweet chestnut (Castanea sativa) is a medium-large monoecious tree that is able to reach 30–35 m and has long-living (up to 1000 years), it is an endemic species of genus Castanea in Europe and is widely distributed in all Mediterranean countries (Fineschi et al. Citation2000; Conedera et al. Citation2016). Villani et al. (Citation1991) showed that the genetic diversity in Turkey deme was greater than that in Italy and France, indicating that Turkey was the secondary origin and genetic diversity center of C. sativa. Some scholars thought that genus Castanea originated in Asian continent (Huang et al. Citation1994), and resulted in the C. sativa by westward migration (Zohary and Hopf Citation1983). Recent research found that Last Glacial Maximum (LGM) refugia of C. sativa were in the north of the Italian, Balkan Peninsulas, Iberian, and Anatolia, and the species spread naturally during the early-middle Holocene (Krebs et al. Citation2019). Roces-Díaz et al. (Citation2017) showed a postglacial expansion of C. sativa in mid-Holocene at more favorable climatic conditions. At present, the complete chloroplast genomes of Castanea seguinii, Castanea henryi, and Castaneamollissima were all reported, however, the complete chloroplast genome of C. sativa is unknown. In this study, we annotated the chloroplast genome of C. sativa into GenBank public database with the accession MW044606.
The leaves of C. sativa were sampled from Taian chestnut garden of national fruit tree germplasm, Shandong province, China. The samples were kept in −80 °C refrigerator at Shandong Institute of Pomology, Taian, China. Genomic DNA of C. sativa leaves was extracted according to CTAB method (Doyle and Doyle Citation1987). The genomic DNA was performed a series of processing according to the standard protocol provided by Illumina company, including genomic DNA purification, library construction, library-quality detection, and paired-end sequencing on Illumina NovaSeq platform. The clean data were obtained from raw sequence data using trim_galore version 0.4.4 software (Babraham Institute, United Kingdom of Great Britain and Northern Ireland) and was aligned to genome of C. mollissima chloroplast (GenBank accession HQ336406) using bowtie2 version 2.2.4 software (Johns Hopkins University, America). The chloroplast genome was assembled by SPAdes version 3.10.1 software (St Petersburg State University, Russia), and all of the genes were annotated using GeSeq through comparison with the genome of C. mollissima chloroplast. The MISA version 1.0 software (MIcroSAtellite identification tool, http://pgrc.ipk-gatersleben.de/misa/misa.html) was used for analysis of simple sequence repeats (SSRs).
The length of chloroplast genome in C. sativa was 160,938 bp. It was a quadripartite structure including a 90,519 bp large single-copy and an 18,967 bp small single-copy, which were separated by a pair of 25,726 bp inverted regions. The total GC content of chloroplast genome in C. sativa was 36.8%. There were 129 genes, including 37 transfer RNA genes, 8 ribosomal RNA genes, and 84 protein-coding genes. In addition, the numbers of SSR were respectively 187 in single base repeat, 17 in double base repeats, 79 in triple base repeats, 11 in quad base repeats, 4 in five base repeats, and 1 in six base repeats.
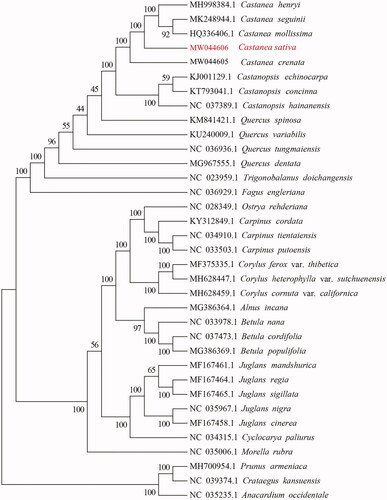
In order to study the molecular phylogenetic relationship, the 35 chloroplast genomes of different species were selected to construct a neighbor-joining (NJ) tree using MEGA software version 6.0 (Auckland, New Zealand) and bootstrap analysis of 1000 replicates (). The main species displayed on the phylogenetic tree were Fagaceae, Betulaceae, and Juglandaceae. In Fagaceae, there was a closer relationship between genus Castanea and genus Castanopsis. Further analysis indicated that C. sativa exhibited the closest relationship with C. henryi.
Disclosure statement
The authors declare that they have no conflict of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW044606. The associated SRA number is SRR13649373.
Additional information
Funding
References
- Conedera M, Tinner W, Krebs P, de Rigo D, Caudullo G. 2016. Castanea sativa in Europe: distribution, habitat, usage and threats. European atlas of forest tree species. Luxembourg: Publication Office of European Union; p. 78–79.
- Doyle JJ, Doyle JD. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fineschi S, Taurchini D, Villani F, Vendramin GG. 2000. Chloroplast DNA polymorphism reveals little geographical structure in Castanea sativa Mill. (Fagaceae) throughout southern European countries. Mol Ecol. 9(10):1495–1503.
- Huang H, Dane F, Norton JD. 1994. Allozyme diversity in Chinese, Seguin and American chestnut (Castanea spp.). Theor Appl Genet. 88(8):981–985.
- Krebs P, Pezzatti GB, Beffa G, Tinner W, Conedera M. 2019. Revising the sweet chestnut (Castanea sativa Mill.) refugia history of the last glacial period with extended pollen and macrofossil evidence. Quat Sci Rev. 206:111–128.
- Roces-Díaz JV, Jiménez-Alfaro B, Chytrý M, Díaz-Varela ER, Álvarez-Álvarez P. 2017. Glacial refugia and mid-Holocene expansion delineate the current distribution of Castanea sativa in Europe. Palaeogeogr Palaeoclimatol Palaeoecol. 491:152–160.
- Villani F, Pigliucci M, Benedettelli S, Cherubini M. 1991. Genetic differentiation among Turkish chestnut (Castanea Sativa Mill.) populations. Heredity. 66(1):131–136.
- Zohary D, Hopf M. 1993. Domestication of plants in the old world. J Appl Ecol, 26(1). DOI: https://doi.org/10.2307/2403680