Abstract
Primula wilsonii Dunn is a perennial herb in section Proliferae Pax of Primula L. with small population sizes in the field. Here, we constructed the complete plastome of the P. wilsonii using Illumina sequencing technology. The circular plastome was 151,677 bp in size, and comprises a large single-copy (LSC) region of 83,510 bp, a small single-copy (SSC) region of 17,765 bp, and a pair of inverted repeats (IR) of 25,201 bp. The GC content was 36.99% overall, with 34.89%, 30.18%, and 42.87% for the LSC, SSC, and IR regions, respectively. The plastome comprised 130 unique genes including 84 protein-coding genes, 37 tRNAs, and 8 rRNAs. The ML phylogenetic analysis based on 17 plastomes in Primulaceae showed a strong sister relationship with P. anisodora in section Proliferae.
Primula L. is a genus of flowering plants with a heterostylous breeding system and extreme species richness, particularly in the eastern Sino-Himalaya region (Richards Citation2003). The Primula species are of high ornamental value, and are famous garden ornamental flower plants. P. wilsonii Dunn is a perennial herb in section Proliferae Pax of Primula (Primulaceae), which is considered as a well-delimited group characterized by numerous whorls of flowers resembling candelabra (Hu Citation1990). Unlike the most other candelabra primulas that are common in the wild and gardon, P. wilsonii is distributed in Sichuan and Yunnan provinces with small population sizes based on our investigation. Here, we reported plastome of P. wilsonii for understanding its systematics and provide scientific basis for formulation of conservation strategy in the future.
Fresh leaves of P. wilsonii for total genomic DNA extraction were collected from Wuxuhai, Sichuan Province, China (29.16 N, 101.41E). The voucher specimen (voucher accession number XYP202007016) was stored at the Key Laboratory of Plant Resource and Biology in Huaibei Normal University. The qualified PCR-amplified library was sequenced with the Illumina NovaSeq Tenplatform (Nanjing Genepioneer Biotechnologies Inc., Nanjing, China). The plastome was assembled using the program NOVOPlasty 2.7.2 (Dierckxsens et al. Citation2016). Annotation was performed using GeSeq (Tillich et al. Citation2017), followed by manual correction for start and stop codons of protein-coding genes. The assembled complete plastome sequence of P. wilsonii was submitted to NCBI, and the accession number is MW442886.
The complete plastome of P. wilsonii was 151,677 bp, consisting of a large single-copy (LSC) region of 83,510 bp, a small single-copy (SSC) region of 17,765 bp, and a pair of inverted repeats (IR) of 25,201 bp. The GC content was 36.99% overall, with unevenly distribution across regions of the plastome, which were found to be 34.89%, 30.18%, and 42.87% for the LSC, SSC, and IR regions, respectively. The plastome comprised 130 unique genes including 84 proteincoding genes, 37 tRNAs, and 8 rRNAs. Nine genes (ndhA, ndhB, petB, petD, atpF, rpl16, rpl2, rps16, rpoC1) contained only one intron and two genes (rps12 and clpP) contained two introns. Two protein-coding genes, seven tRNAs, and all 8 rRNAs were completely duplicated within IRs.
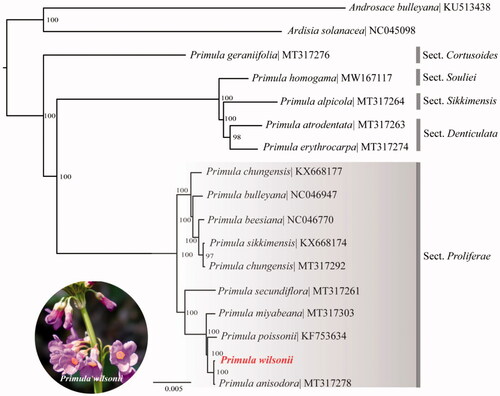
To further investigate its phylogenetic position in Primulaceae, especially in section Proliferae, a maximum likelihood tree was constructed based on complete plastome sequences of 17 species in Primulaceae by online RAxML BlackBox software (Stamatakis et al. Citation2008), after the sequences were aligned using MAFFT v7.307 (Katoh and Standley Citation2013). Our results suggested P. wilsonii was close to the other candelabra primulas species, especially closer to P. miyabeana, P. poissonii and the sister species P. anisodora (). The cluster including these species are accordant with the results of previous phylogenetic studies of Primula (Yan et al. Citation2011, Citation2015). This published P. wilsonii plastome might provide useful information for phylogenetic and evolutionary studies in section Proliferae and Primulaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete plastome sequence and annotation of Primula wilsonii that support the findings of this study are openly available in Zenodo ((doi: 10.5281/zenodo.4435975) at https://zenodo.org/record/4435975#.X_6ybjnivic.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Hu CM. 1990. Primulaceae. In: Chen FW, Hu CM, editors. Flora Reipublicae Popularis Sinicae. Beijing: Science Press; Vol. 59; p. 118–119.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Richards J. 2003. Primula (new edition). Oregon: Timber Press.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 57(5):758–771.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Yan HF, Hao G, Hu CM, Ge XJ. 2011. DNA barcoding in closely related species: a case study of Primula L. sect. Proliferae Pax (Primulaceae) in China. J Syst Evol. 49 (3):225–236.
- Yan HF, Liu YJ, Xie XF, Zhang CY, Hu CM, Hao G, Ge XJ. 2015. DNA barcoding evaluation and its taxonomic implications in the species-rich genus Primula L. in China. PLOS One. 10(4):e0122903.